Extended Data Fig. 3. Haplotype copy number and SVs for the targeted chromosome for each sample in the paper.
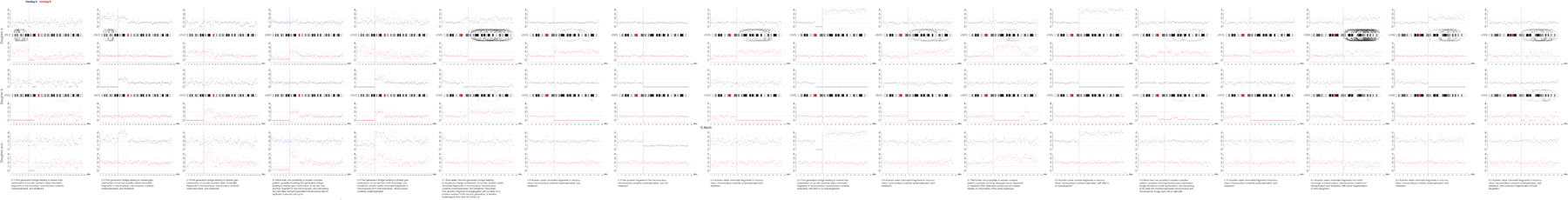
Haplotype-resolved copy number and structural variant analysis for the targeted chromosome for each granddaughter pair. Red and blue dots represent 1 Mb copy number bins for each homolog, and curved lines represent structural variants of ≥ 1 Mb that could be on either homolog. Top, ‘granddaughter a’; middle, ‘granddaughter b’; bottom, sum copy number for each homolog for the pair of cells. Note that in most cases there should be a total of two red and two blue copies per granddaughter pair, and deviation from this represents certain missegregation or events, such as first-generation bridge formation. Copy number alterations occurring only in one daughter without a corresponding or reciprocal change in the other daughter were attributed to random noise due to variability in genome amplification quality. Text: inferred most likely explanation for each copy number and rearrangement profile. Note that alternative explanations exist for many samples, such as a G1 cut followed by replication of the cut chromosome.
