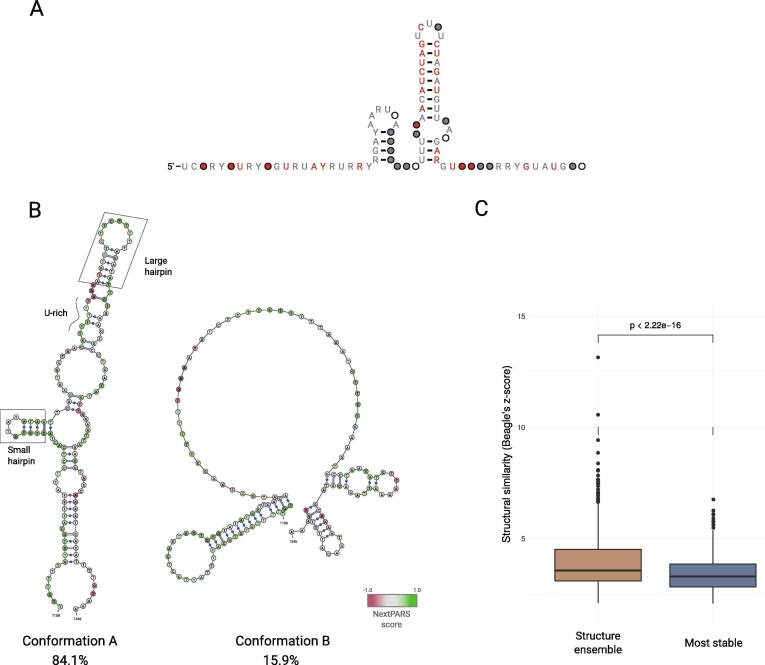
Fig. 4.
Comparison of alternative structures of NRUs and obtained consensus structure. A. The consensus secondary structure for the core region of NRUs, derived by using a multiple sequence alignment to derive a from the eight NRUs that have a clear conserved long stem-loop hairpin (NRU#1, NRU#3–8 and NRU#10). B. Secondary structure conformations A and B, from NRU#2 obtained using Rsample software in combination with nextPARS from larger NORAD fragments experiments. Scores from nextPARS scores are depicted by colored nucleotides. Known structural and sequence motifs are marked in the conformation A. C. A box plot showing the structural similarity (Beagle's z-score) elements found by Beagle software, for the structure ensemble alignment and for the most stable structure alignment.