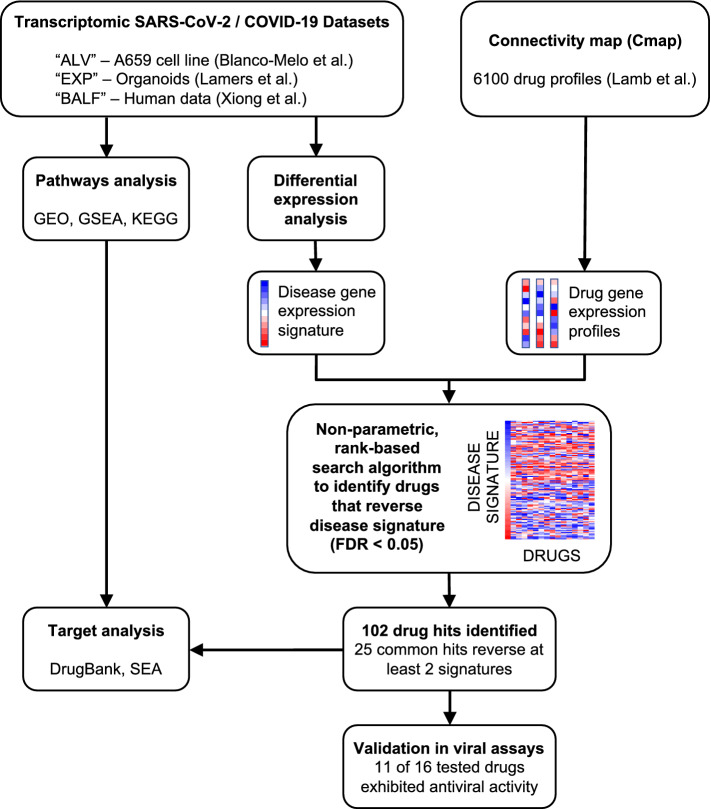
Figure 1.
COVID-19 transcriptomics-based bioinformatics approach for drug repositioning. We generated lists of statistically significant differentially expressed genes from the analysis of three published studies of SARS-CoV-2 and COVID-19. The drug repositioning computational pipeline compares the ranked differential expression of the COVID-19 disease signature with that of drug profiles from CMap. A reversal score based on the Kolmogorov–Smirnov statistic is generated for each disease-drug pair. If a drug profile significantly (FDR < 0.05) reverses the disease signature, then the drug could be therapeutic for the disease. Across all datasets, a total of 102 drugs have been identified as potentially therapeutic for COVID-19. Twenty-five drugs were identified in analyses of at least two of the three datasets. We further conducted pathways analyses and targeted analyses on the results, focusing on the 25 shared hits. Finally, we validated 16 of our top predicted hits in live SARS-CoV-2 antiviral assays.