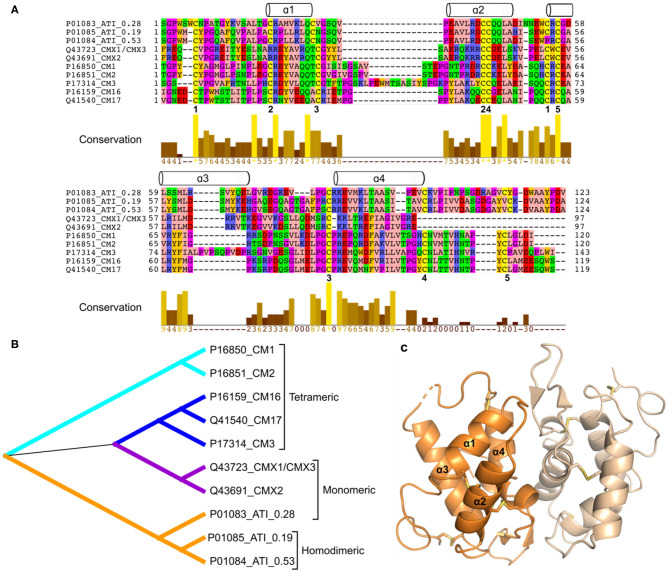
Figure 2.
(A), Multiple sequence alignment including wheat ATIs (signal peptides have been removed). The wheat ATIs only share very few fully conserved positions including some of the disulphide bonds (numbered 1–5 below the alignment). The α-helices above the alignment indicate the secondary structure elements based on the 3D structure of ATI 0.19 (part C), and the alignment is colored according to amino acid properties. (B), Phylogenetic tree generated based on alignment shown in (A). (C), Three-dimensional structure of wheat homodimeric ATI 0.19 [PDB entry 1HSS (11)]. Although only one 3D structure has been determined for 0.19, and the sequence identity is low, all wheat ATIs are predicted to share the following overall structure: four α-helices connected by irregular loop regions and stabilized by disulphide bonds (a four-α-helix bundle). Software used: MEGA X (12) (Muscle for alignment preparation and Maximum likelihood for phylogenetic analysis), Dendroscope (13), Jalview (14), and PyMOL (Schrödinger, LLC).