FIGURE 1.
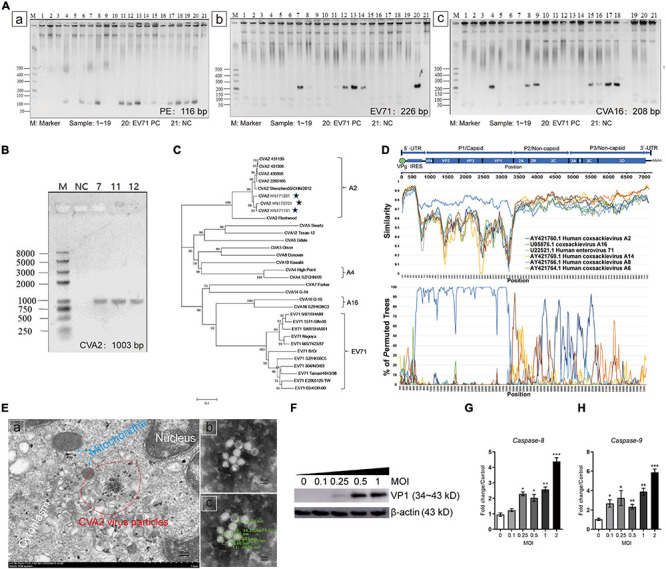
Identification of CVA2 strains in clinical isolates from HFMD patients. (A) PCR analysis of 19 patient samples. a. samples 4, 6, 7, 8, 9, 11, 12, 13, 14, 15, 16, 17, 18, and 19 showed positive PE nucleic acid; b. Samples 7, 8, 11, 12, 13, and 14 were positive for EV71 nucleic acid; and c. Samples 4, 6, 8, 9, 15, 16, 17, and 18 were positive for CVA16 nucleic acid. (B) The PCR products (1,003 bp) of samples 7, 11, and 12. (C) A genetic evolutionary tree of samples 7, 11, and 12 VP1 (1,003 bp) by using the neighbor-joining method. (D) Recombination analysis of the full-length genome of HN202009 by using the Simplot. (E) Transmission electron micrograph of the infected Vero cells and inactivated CVA2 virions. a. Virus inclusion bodies in infected Vero cells; b and c. Spherical particles of the CVA2 virions with a diameter of 26∼35 nm. (F) The relative expression of VP1; and (G,H). The expression of caspase-8 and caspase-9 RNA in the infected cells with increased doses of CVA2. Positive control, PC; negative control, NC. *P < 0.05; **P < 0.01; ***P < 0.001.
