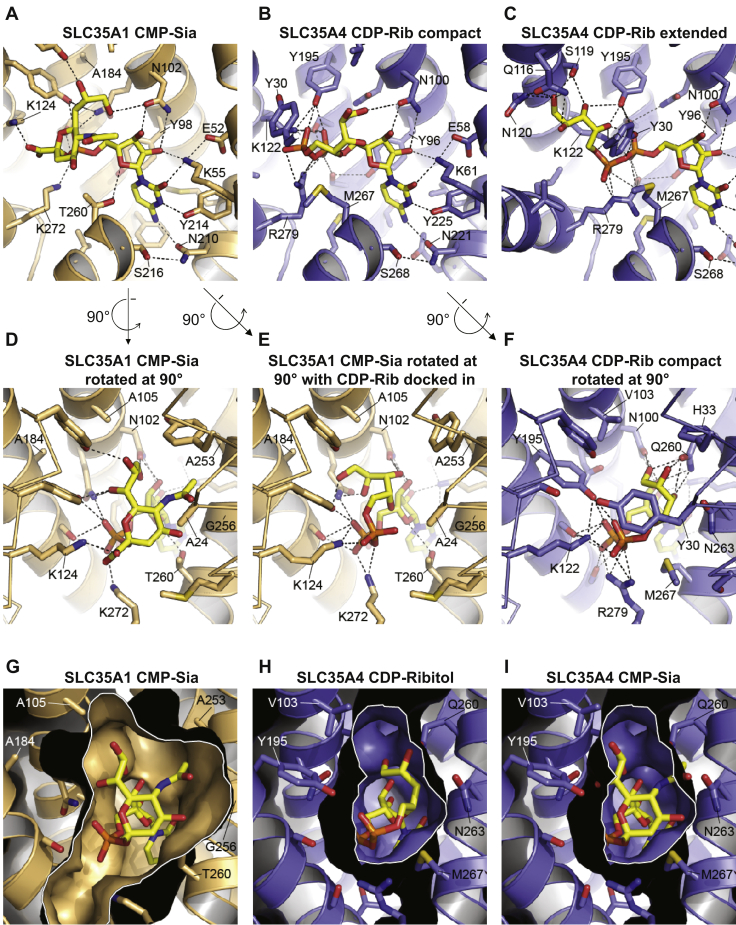
Figure 5.
Comparison of substrate-binding sites in SLC35A1 and SLC35A4.A, the structure of CST in complex with CMP-Sia (yellow molecule) is shown. B and C, the structural homology model of SLC35A4 is shown with CDP-ribitol (yellow molecule) computationally docked in either the “compact” (B) or “extended” (C) conformation. In panels A–C, portions of transmembranes (TMs) 1 and 8, which are in front of the substrate, are hidden for clarity. D, a rotated view of the structure of CST in complex with CMP-Sia shown in panel A. E, the same view of the CST structure as in panel D, but with CDP-ribitol computationally docked in. F, a rotated view of what is shown in panel B: the model of SLC35A4 with CDP-ribitol in the compact conformation. In panels D–F, Cα traces are shown for TMs 1 and 7 (instead of cartoon ribbons) for clarity. In panels A–F, key residues are indicated, and select polar interactions are indicated by dashed lines as a visual aid. G, in a view that is similar to that shown in panel D, a surface representation is shown for the CST structure in complex with CMP-Sia. H and I, in a view that is similar to that shown in panel F, a surface representation is shown for the model of SLC35A4 with CDP-ribitol in the compact conformation (H) or with CMP-Sia manually placed into the cavity to show the clashes that would occur between the Sia moiety and several cavity-lining side chains. In panels G–I, a white border outlines the sliced view through the substrate-binding cavity. In all panels, only side chains that are within 5 Å of the substrate are shown for clarity. CMP-Sia, CMP-sialic acid; CST, CMP-sialic acid transporter.