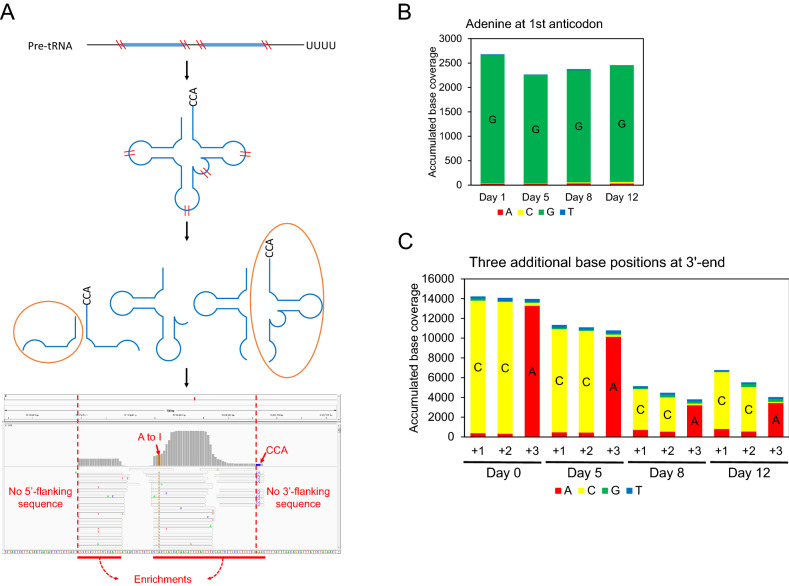
Figure 1.
Characteristics of tRFs identified by sequencing read alignment to the genome reference. (A) Example of sequencing read alignment to a tRNA gene locus is shown with a brief illustration of tRF generation via tRNA maturation. The sequencing read alignment suggests that the majority of tRFs are derived from mature tRNAs. Upon transcription, pre-tRNA has 5′ and 3′ margins as well as an intron, which are removed later with the maturation process. In addition, 3′-CCA and RNA base modifications observed in the sequencing reads are characteristics of mature tRNA. Dashed vertical lines at the integrative genomics viewer (IGV) plot indicate the start and the end of the tRNA gene. (B) Accumulated base coverage at the first position of the anticodons starting with adenine. The base coverage for every age is shown as a bar graph. For 56 first anticodon base loci, for which the genomic sequence is adenine, the sequenced bases were counted per read alignment. (C) Accumulated base coverage at the three additional base positions of all tRNA genes. For the three additional 3′ base positions of 600 tRNA genes (excluding pseudogenes), the sequenced bases were counted per read alignment. For both panels (B) and (C), adenine (A), cytosine (C), guanine (G), and thymine (T) bases are indicated by red, yellow, green, and blue colors, respectively. The height of colored bar indicates the number of sequencing reads with a corresponding base at each locus.