Abstract
CD4+ T cells orchestrate adaptive immune responses via binding of antigens to their receptors through specific peptide/MHC-II complexes. To study these responses, it is essential to identify protein-derived MHC-II peptide ligands that constitute epitopes for T cell recognition. However, generating cells expressing single MHC-II alleles and isolating these proteins for use in peptide elution or binding studies is time consuming. Here, we express human MHC alleles (HLA-DR4 and HLA-DQ6) as native, noncovalent αβ dimers on yeast cells for direct flow cytometry-based screening of peptide ligands from selected antigens. We demonstrate rapid, accurate identification of DQ6 ligands from pre-pro-hypocretin, a narcolepsy-related immunogenic target. We also identify 20 DR4-binding SARS-CoV-2 spike peptides homologous to SARS-CoV-1 epitopes, and one spike peptide overlapping with the reported SARS-CoV-2 epitope recognized by CD4+ T cells from unexposed individuals carrying DR4 subtypes. Our method is optimized for immediate application upon the emergence of novel pathogens.
Keywords: yeast display, MHC ligands, T cell epitopes, RIPPA, SARS-CoV-2 S
Subject terms: MHC class II, Antigen presentation
Introduction
CD4+ T cell responses are crucial drivers of defensive immunity against infection; however, they can also cause autoimmune responses when tolerance is broken. These T cells respond to antigens via interactions between T cell receptors (TCRs) and antigen-derived peptides bound to heterodimeric (α/β) major histocompatibility complex class II (MHC-II) molecules on the surface of professional antigen-presenting cells (APCs) [1, 2]. Identification of T cell responses specific for a target antigen often requires intensive screening of overlapping peptides covering the candidate protein in a T cell assay. The use of characterized peptide/MHC complexes as probes for TCR binding or TCR-mediated T cell activation allows further assessment of reactive T cell clones. For example, we and others have applied both in vitro T cell assays and ex vivo peptide/MHC-II probes to identify CD4+ T cell clones targeting a neurotransmitter (hypocretin, HCRT) in narcolepsy patients [3–5]. Recently, similar analyses have identified CD4+ T cell responses to SARS-CoV-2 viral proteins [6–9]; however, a lack of well-characterized MHC-II ligands derived from these viral antigens limits the examination of reactive T cell clones.
The T cell repertoire is tremendously diverse, and TCRs expressed by distinct clones have different specificities for particular peptides bound to particular MHC-II allelic proteins [10, 11]. Thanks to the development of MHC (class I and II) tetramer staining technology [12], one can stain and isolate peptide/MHC tetramer-positive T cell clones for functional and structural investigations [13–15]. The current limitation is that tetramer synthesis requires information on the binding of MHC molecules to peptides derived from the candidate protein antigen. Both computational and experimental efforts have been made to generate such binding information, with the former method relying largely on empirical data [16–20]. There are two major experimental approaches to identify MHC-II ligands. One uses mass spectrometry to quantify peptides eluted from MHC-II molecules that are immunoprecipitated from lysed cells [20–23]. The other detects the binding of synthesized peptides to soluble, recombinant MHC-II proteins [5, 13, 24–27]. Currently, to identify peptides bound by a particular MHC-II allelic protein via either method, it is optimal to generate cell lines that express only this allele, as primary cells in humans are typically heterozygous and codominantly express both alleles from each of the HLA-DR, HLA-DQ, and HLA-DP genes that encode the three isotypes of human leukocyte antigen (HLA). In addition, both methods require purification of the expressed MHC-II protein. Based on our previous experience [5, 24, 28], these steps typically take up to 4 months, significantly limiting the speed of empirical studies.
Here, we develop a new methodology that allows elimination of the labor-intensive expression/purification steps by taking advantage of yeast display [29]. As single-cell eukaryotes, yeast cells have the fast cloning capability of Escherichia coli and are equipped with posttranslational modification machinery similar to that in mammalian cells [29, 30]. Therefore, linking an exogenous protein (e.g., MHC-II) to a native yeast protein on the yeast cell surface offers a fast way to investigate the function of the exogenous protein. To express MHC-II alleles, including DR and DQ, as noncovalent heterodimers without an interchain linker on the yeast cell surface, we replace the transmembrane domains with leucine zipper (LZ) dimerization motifs [31, 32]; this LZ fusion facilitates pairing of α/β chains that are encoded by a bidirectional expression construct and secreted separately by yeast cells [33, 34]. We prove that both the DR and DQ constructs are correctly folded without the necessity for covalently linked peptides and are functional in binding exogenous peptides. We then design a competition assay that enables rapid identification of MHC-II peptide ligands from protein antigens (RIPPA), using HCRT and the SARS-CoV-2 spike (S) as model antigens. The quick setup time (<1 month) for the RIPPA in vitro peptide-binding assay allows efficient testing of MHC-II ligands to guide tetramer synthesis and expedite downstream investigation of cell-mediated immune responses that are relevant to disease. These characteristics are particularly useful in the setting of novel, rapidly spreading diseases, such as COVID-19.
Results
Leucine zippers enhance MHC-II expression on yeast cells independent of peptide ligands
Professional APCs express MHC-II molecules as α/β heterodimeric membrane proteins initially associated with the chaperone invariant chain (Ii) in the endoplasmic reticulum. The peptide-binding groove of nascent MHC-II is occupied by a region of Ii that is proteolytically trimmed via the endosomal pathway to yield the CLIP peptide [35]. An antigenic peptide capable of binding to a given MHC-II allelic protein can replace CLIP through a peptide exchange process. This process most often takes place in the late endosomal MHC-II-enriched compartment (MIIC), where it is typically catalyzed by HLA-DM [36], although cell surface exchange may occur under some conditions [37]. It is likely that mass spectrometry underestimates the abundance of certain MHC-II binders in the eluted ligandome, when these binders are outcompeted by highly abundant or high-affinity peptides due to physiologic (e.g., intracellular cleavage or DM effects) or experimental (e.g., differences between model cell lines and primary cells) conditions. Therefore, to explore all possible MHC-II ligands from candidate antigens, it is ideal to evaluate MHC-II binding of overlapping peptides spanning the entire antigen. When recombinant MHC-II ectodomains are used in binding studies, a specific peptide ligand is typically linked to the N terminus of the MHC-II β chain to stabilize the α/β dimers. A linker cleavage step is then necessary to ensure the production of an exchangeable placeholder peptide for a peptide-binding assay [24].
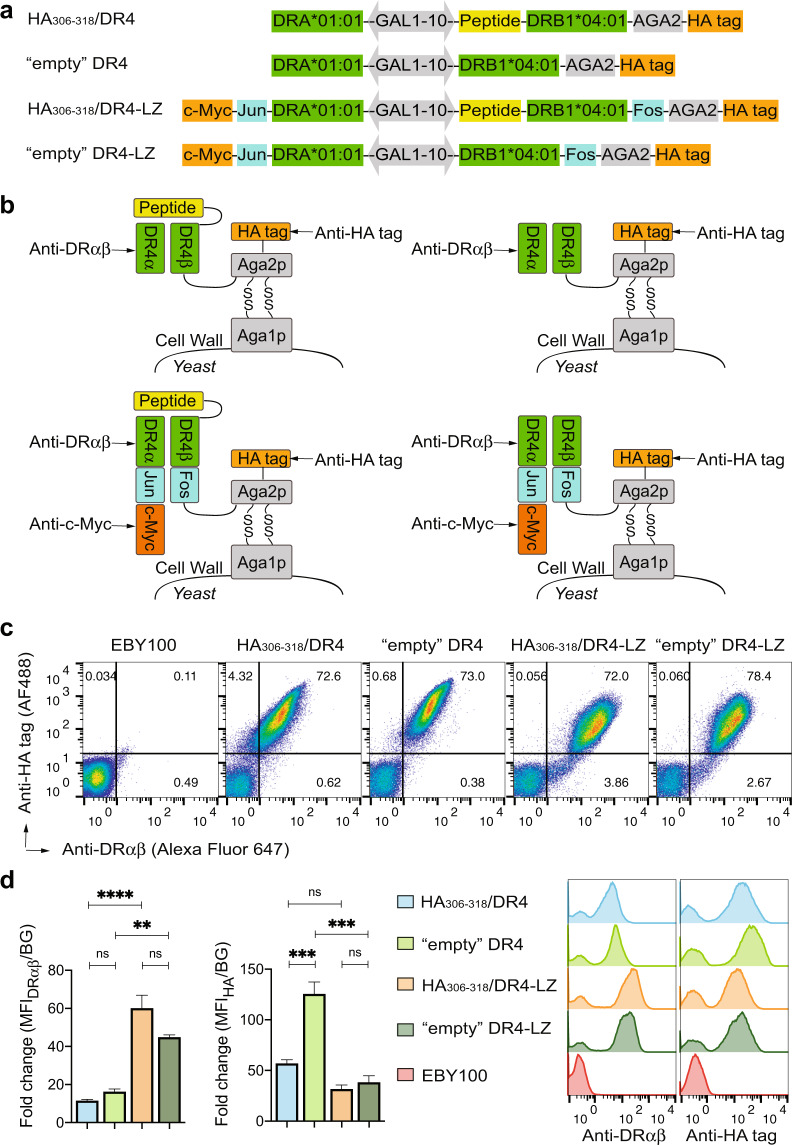
To avoid the requirement for the linker cleavage step in our peptide-binding assay using yeast display of MHC-II, we first utilized an “empty” construct expressed by yeast. We used DR4 (DRA*01:01/DRB1*04:01) as a representative DR allele, along with an influenza hemagglutinin (HA)306–318 peptide-linked construct that was previously examined in yeast [34]. Importantly, a bidirectional GAL1–10 promoter was used to direct simultaneous expression of the α and β chains from a yeast shuttle vector (Fig. 1a). Unlike the single-chain format of recombinant DR proteins used in several previous attempts at yeast display [16, 38–41], the noncovalent native format no longer requires mutation of MHC-II to facilitate protein folding. Considering the potential instability of the “empty” DR4 protein, we included LZ motifs [31, 32] in two additional constructs to facilitate dimerization (Fig. 1a). Previously, LZ Fos/Jun motifs were used to allow α/β pairing of a single-chain DR protein in yeast, although the resulting protein was not functional for peptide binding [39]. In all four constructs developed herein, the DR4 β chain ectodomain with or without the Fos motif was engineered to be located upstream of the yeast AGA2 gene and followed by an HA epitope tag, while the α chain ectodomain with or without the Jun motif was designed to be secreted from yeast (Fig. 1a). Successful folding and α/β pairing can yield a functional DR4 protein expressed as a fusion to the yeast native surface protein agglutinin, which is composed of the Aga1p and Aga2p subunits (Fig. 1b).
Fig. 1. The leucine zipper motif enhanced proper folding and surface expression of functional DR4 in yeast.
a Gene construct used for yeast display of the noncovalent DR4 ectodomain with or without covalently linked HA306–318, a hemagglutinin-derived peptide. The bidirectional GAL1–10 promoter directed the expression of the α and β chains. The Fos/Jun LZ motifs in the last two constructs were used to facilitate α/β pairing. b Schematic illustration of appropriately assembled HA306–318/DR4 or “empty” DR4 as a fusion to the yeast surface protein agglutinin, composed of the Aga1p and Aga2p subunits. The arrows indicate protein or epitope tags for antibody staining and detection by flow cytometry. c Expression of HA306–318/DR4 and “empty” DR4 on the surface of yeast, as analyzed by flow cytometry. Yeast cells transformed with one of the constructs described in a were induced to express proteins and were double-stained with anti-DRαβ (clone L243) and anti-HA-tag antibodies. Untransformed yeast cells (EBY100) were used for background staining. d Comparison of DR4 expression levels in different yeast transformants. The fold change in the median fluorescence intensity (MFI) of DRαβ or HA-tag signals on the surface of transformants with respect to the background (BG) fluorescence signal was quantified. Representative histograms are shown to the right. The error bars indicate the standard error of the mean (SEM) of at least three independent experiments. Significance was determined using one-way ANOVA. ns: p > 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
We generated four yeast strains by individually transforming the above constructs into the parent Saccharomyces cerevisiae strain EBY100 [29]. After induction of protein expression, we detected properly assembled DR4 on the surface of all four strains by costaining with two antibodies, one specific for DRαβ (clone L243) and the other specific for the HA tag (Fig. 1c). As expected, the LZ enhanced the surface level of folded DR4 by 5× and 3× for the peptide-linked and “empty” constructs, respectively, as measured by flow cytometry (Fig. 1d). This improvement was attributed to α/β dimerization facilitated by the LZ and not to yeast protein production, as the level of Aga2p expression, indicated by staining of the HA tag, were not correlated with the levels of folded dimers (Fig. 1d). Notably, as L243 is typically specific for stable DR4 heterodimers in association with peptides, the similar levels of “empty” and peptide-linked DR4 stained by L243 implied that the “empty” binding grooves were likely occupied by peptides derived from the cultured yeast cells [41] rather than being truly empty (herein, quotation marks are used to indicate that the constructs were genetically empty). To further confirm that both chains of DR4 with the LZ motifs were expressed by yeast, we simultaneously stained the α and β chains. Costaining of the c-Myc-tag at the C terminus of the α chain (Fig. 1a) and the HA-tag at the C terminus of the DRβ-Aga2p fusion confirmed the expression of both the α- and β-encoding open reading frames (Supplementary Fig. 1). The presence of both chains on the surface was also consistent with the detection of correctly folded DRαβ by the L243 antibody (Fig. 1c).
Yeast display of peptide-linked or “empty” DQ molecules as noncovalent heterodimers
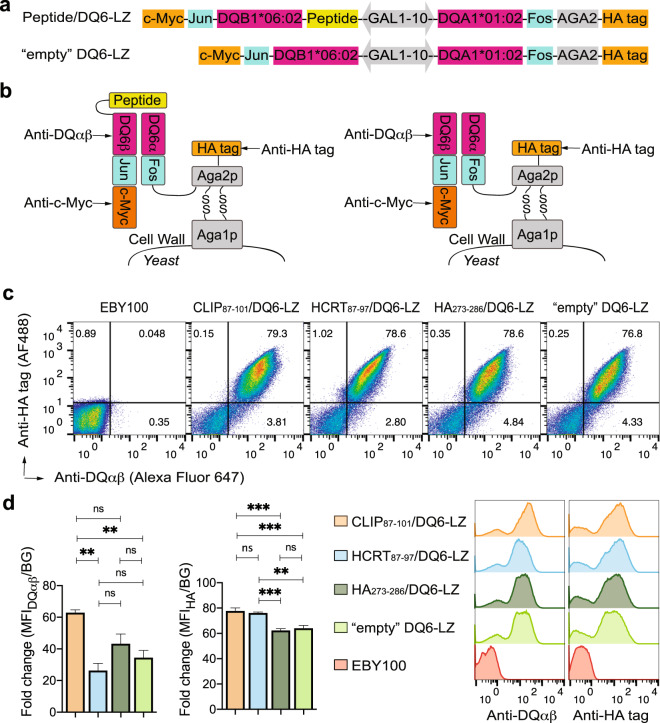
Next, we tested the expression of a representative DQ allele, DQ6 (DQA1*01:02/DQB1*06:02), on the yeast surface via the same strategy used to evaluate DR4 except with the β chain as the secreted component (Fig. 2a). Successful chain pairing enabled surface display of a functional DQ6 ectodomain (Fig. 2b). It is known that individuals carrying this DQ6 allele are susceptible to narcolepsy [3–5], and during the 2009 flu pandemic, a significant increase in the narcolepsy incidence occurred among DQ6+ populations in association with natural infection or a particular vaccine formulation [42, 43]. Several reports have identified DQ6-binding antigenic peptides in both the self-protein HCRT and viral proteins, including HA, expressed by the 2009 H1N1 influenza virus [4, 5]. In the peptide-linked constructs (Fig. 2a), we included the Ii peptide, CLIP87–101, and two other peptides, i.e., HCRT87–97 and H1N1-HA273–286, whose binding to DQ6 has been characterized [4, 5, 44].
Fig. 2. Proper folding and surface expression of peptide-linked or “empty” DQ6 with a LZ domain in yeast.
a Gene construct used for expression of the noncovalent DQ6 ectodomain with or without specific peptides covalently linked to the DQ6β N terminus. In the peptide-linked constructs, we included two peptides, HCRT87–97 and HA273–286, whose binding to DQ6 has been well characterized, and a positive control Ii peptide, CLIP87–101. Unlike in the DR construct, the α chain upstream of the Fos LZ motif was fused directly to the N terminus of the Aga2p subunit. b Schematic illustration of appropriately assembled peptide-linked or “empty” DQ6 as a fusion to the yeast surface protein agglutinin (similar to Fig. 1b). c Expression of peptide/DQ6 or “empty” DQ6 on the surface of yeast, as analyzed by flow cytometry (as in Fig. 1c, except that an anti-DQαβ antibody (clone SPV-L3) was used). d Comparison of DQ6 expression levels in different yeast transformants. The fold change in MFI with respect to the background (BG) fluorescence signal was quantified (as in Fig. 1d). Representative histograms are shown to the right. The error bars indicate the SEM of at least three independent experiments. Significance was determined using one-way ANOVA. ns: p > 0.05, **p < 0.01, ***p < 0.001.
After gene transformation and protein induction, similar to the DR4-expressing strains, all four yeast strains expressing the DQ6 constructs showed a double-positive population upon surface costaining with antibodies specific for the HA tag and a DQαβ conformational determinant detected by the monoclonal antibody (mAb) clone SPV-L3 (Fig. 2c). Notably, the level of appropriately folded DQαβ varied significantly, with the linked CLIP87–101-linked construct having the highest level (Fig. 2d). Costaining of the c-Myc-tag and the HA-tag, as performed to evaluate DR4 expression, further confirmed the expression of both the α and β ectodomains of DQ6 in yeast (Supplementary Fig. 2). The relative expression level of the secreted β chain, indicated by the fold change in c-Myc-tag staining, was significantly higher for the CLIP87–101/DQ6-LZ construct, consistent with the fold change in DQαβ staining.
Collectively, both the “empty” DR and DQ constructs yielded properly assembled MHC-IIαβ on the surface of yeast cells, facilitated by LZ dimerization. Although the presence of linked peptides may influence protein expression, the successful display of “empty” MHC-II as a noncovalent heterodimer enabled further evaluation of the capacity of MHC-II molecules to accommodate exogenously added peptides.
“Empty” MHC-II on yeast binds specific peptides under various conditions
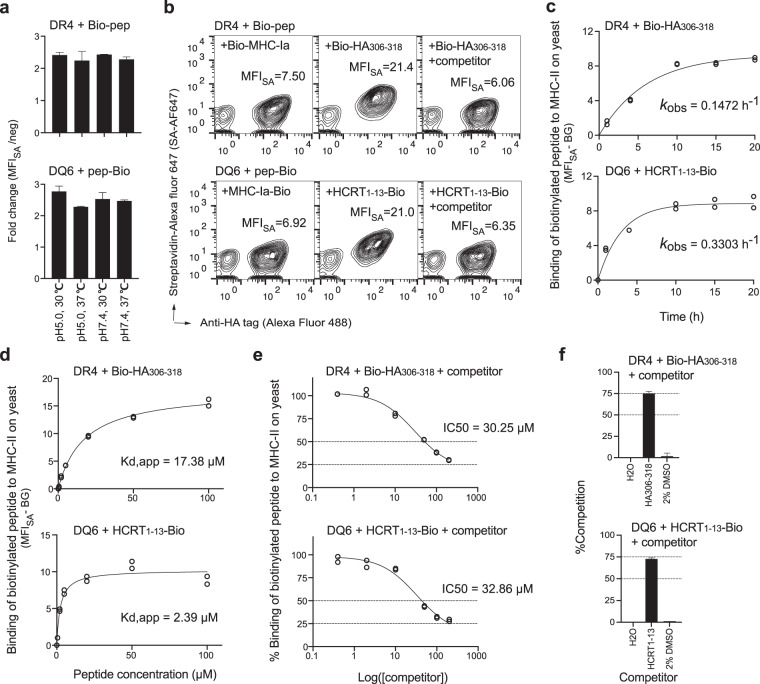
Physiologic peptide loading occurs in the acidic MIIC (pH ~5) at human body temperature (37 °C) [35]. Yeast cells express proteins at 30 °C in culture systems with pH values ranging from 5 to 7. To evaluate yeast display as a robust system to study MHC-II peptide binding, we examined various binding conditions. Yeast incubated with an indicator biotinylated peptide ligand consistently yielded 2–3× higher biotin signals than yeast incubated with the irrelevant biotinylated peptide control at pH 5.0 vs pH 7.4 and at 30 °C vs 37 °C (Fig. 3a, b and Supplementary Fig. 3a). The capacity to bind exogenous peptides validated the functional integrity of noncovalent MHC-II α/β heterodimers on yeast. Incubation conditions of pH 5.0 (physiological binding pH) and 30 °C (habitual temperature for yeast survival) were then used in subsequent experiments.
Fig. 3. Binding of peptides to “empty” MHC-II displayed on yeast.
a Yeast cells expressing “empty” MHC-II were incubated with 20 μM biotinylated peptides (Bio-HA306–318 for DR4 or HCRT1–13-Bio for DQ6) in different pH buffers at the indicated temperature for 20 h prior to streptavidin (SA)-Alexa Fluor 647 staining and flow cytometric analysis. The fold change in MFISA with respect to the negative control (an irrelevant MHC-Ia peptide) was quantified. b Representative flow cytometric measurement of the MFISA of biotinylated peptides bound by MHC-II. The right panel shows data for cultures containing 200 μM nonbiotinylated ligands (HA306–318 for DR4 or HCRT85–99 for DQ6) as competitors. Incubation conditions (b–f): pH 5.0, 30 °C, 20 h (unless otherwise indicated). c Yeast cells were incubated with 20 μM biotinylated peptides for various durations. The BG-subtracted binding signals were plotted against time and fitted to calculate the observed rate constant (kobs). d Yeast cells were incubated with different concentrations of biotinylated peptides for 20 h to reach equilibrium. Data approximating equilibrium binding at each peptide concentration were fitted to calculate the apparent equilibrium dissociation constant (Kd,app). e Yeast cells were incubated with 20 μM biotinylated peptides and various concentrations of competitor peptides (HA306–318 for DR4 or HCRT1–13 for DQ6). The equation %Binding = (MFIwith competitor − BG)/(MFIwithout competitor − BG) × 100%, was fitted to calculate IC50 values. f Yeast cells were incubated with 20 μM biotinylated and 200 nonbiotinylated peptides or negative controls, as indicated. %Competition = 100% − %Binding (calculated as described in e). Tight duplicates from a representative experiment are shown (c–e). All experiments were repeated at least three times with similar results, and the error bars indicate the SEM values (a, f).
Many endpoint approaches, such as the capture ELISA-based peptide loading assay [24, 25, 28], often require >10 h of incubation to reach equilibrium. These approaches rely on competition between competitor peptides and an indicator peptide to characterize relative MHC-II binding capacities, i.e., the half-maximal inhibitory concentration (IC50), rather than absolute kinetic parameters, i.e., on/off rates and dissociation constant (Kd) values. Similarly, the flow cytometry-based yeast display approach was optimized for endpoint comparison of relative binding capacities (Fig. 3b). Notably, given the assumption of a pseudo-first-order reaction (see “Methods”), we can estimate the observed rate constant (kobs) in a time course experiment (Fig. 3c and Supplementary Fig. 3b). Similar to the capture ELISA [24], the yeast display approach suggested kinetics in which equilibrium was reached after 10 h. Using a 20 h incubation time to approximate the equilibrium binding at each peptide concentration, we calculated the apparent dissociation constant (Kd,app) for the binding of peptides to either “empty” MHC-II expressed on yeast (Fig. 3d and Supplementary Fig. 3c) or soluble MHC-II (Supplementary Fig. 3e). Measurements using the yeast display system can be as good as those using soluble proteins, despite noticeable range differences, compared to kinetic approaches that are optimal for quantification of absolute Kd values [27]. Yeast display also differs from other approaches due to the complex microenvironment on the cell surface (nonspecific binding, uneven local distribution, mass-transfer limitation, etc.). However, these differences did not interfere with the determination of relative binding capacities (see below).
Using a concentration (20 μM) of biotinylated indicator peptides that resulted in minimal nonspecific binding (as indicated by the negligible background (BG) staining at ≤20 μM shown in Supplementary Fig. 3c), we observed concentration-dependent inhibition of indicator binding in the presence of its nonbiotinylated counterpart on yeast (Fig. 3e and Supplementary Fig. 3d). The calculated IC50 values (~30 μM) were very close to the theoretical value (20 μM). Importantly, an orthogonal ELISA-based approach using soluble MHC-II validated the yeast display measurements (Supplementary Fig. 3f). To show the relative binding of competitor peptides, we converted the %Binding value corresponding to the indicator to a %Competition (=100% − %Binding) value corresponding to the competitor. Of note, when the competitor and indicator were the same peptide, 50%Competition reflected the theoretical value when [competitor]:[indicator] = 1; ~75%Competition reflected an empirical average when [competitor]:[indicator] = 10 using both soluble and yeast-displayed MHC-II (Fig. 3f and Supplementary Fig. 3g, h). Therefore, we applied two cutoff values (50%Competition and 75%Competition) to estimate how well a tested peptide competitively bound a given MHC-II molecule with reference to the indicator peptide.
The spontaneous peptide loading and competition suggests that “empty” MHC-II molecules on yeast are preloaded with low-affinity peptides that can be easily replaced by exogenous peptides, unlike the conditions for our previous peptide loading of soluble DQ6, which required the addition of soluble DM to catalyze the replacement of prebound CLIP [24]. The competition assay in yeast thus offers a simplified and robust platform for rapid identification of (unknown) MHC-II peptide ligands from a candidate protein antigen (RIPPA).
Similar ligands are identified using yeast display, soluble pMHC-II, and computational prediction approaches
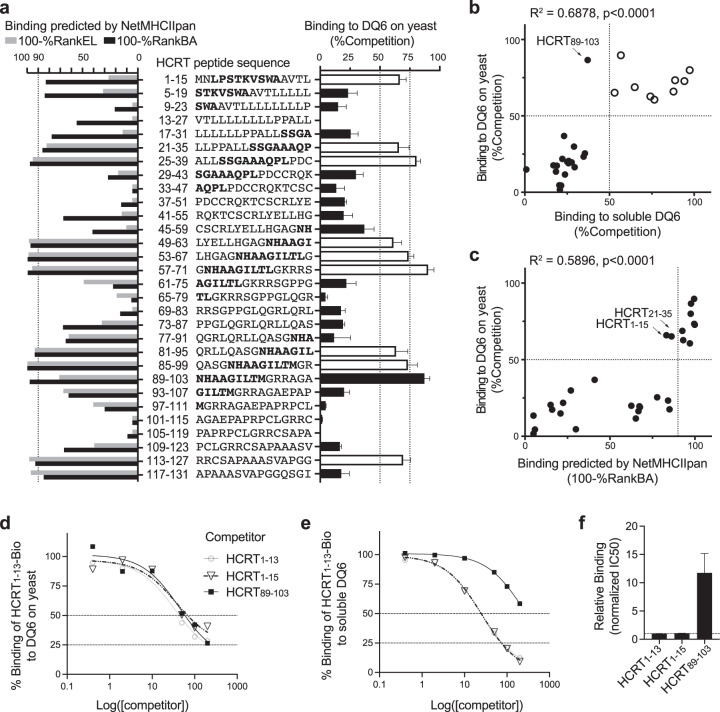
As a demonstration of the yeast-facilitated RIPPA methodology, we screened a set of overlapping 15-mer peptides (Fig. 4a) covering the hypocretin precursor (pre-pro-HCRT) for DQ6 ligands. As reported previously [5], using the same set of overlapping peptides, we identified five regions in the pre-pro-HCRT protein that generate peptides capable of binding to soluble DQ6 protein; we then utilized the corresponding HCRT peptide/DQ6 tetramers to isolate in vivo-expanded CD4+ T cells from narcoleptic patients and healthy controls. Comparison of the peptide-binding data generated using yeast-displayed DQ6 vs that generated with soluble DQ6 [5] vs the computational prediction by the widely used NetMHCIIpan-4.0 server [19] allowed us to examine the efficiency and accuracy of RIPPA (Fig. 4a–c).
Fig. 4. RIPPA identified all DQ6 binders from HCRT.
a Right panel: yeast expressing “empty” DQ6 incubated with 20 μM HCRT1–13-Bio and the indicated nonbiotinylated HCRT 15-mer peptide (200 μM) at pH 5.0 and 30 °C for 20 h and were then analyzed (as in Fig. 3f). The error bars indicate the SEM of four independent experiments. The bolded letters denote previously identified 9-aa DQ6-binding registers [4, 5, 44]: LPSTTKVSWA, SSGAAAQPL, NHAAGILTL, and NHAAGILTM. Left panel: binding ranks of each HCRT peptide predicted by NetMHCIIpan-4.0 shown as 100% − %RankEL or 100% − %RankBA (see “Methods” for details). b, c Correlation analysis of binding data acquired using “empty” DQ6 displayed on yeast vs soluble DQ6 protein (b) or vs NetMHCIIpan-4.0 (BA) predictions (c). The arrows indicate peptides that show binding by one method but not the other in the comparison. The open circles in b match the open bars in a and indicate ligands identified by both empirical methods. d Yeast cells were incubated with 20 μM HCRT1–13-Bio and various concentrations of competitor peptides and were then analyzed (as in Fig. 3e). e Soluble DQ6 (25 nM) was incubated with 20 μM HCRT1–13-Bio and various concentrations of competitor peptides (as in d) in the presence of 100 nM soluble DM. The amount of DQ6-bound HCRT1–13-Bio under each condition was quantified by capture ELISA using europium (Eu) time-resolved fluorescence (see “Methods”). %Binding = (Eu-SAwith competitor − BG)/(Eu-SAno competitor − BG) × 100%. The mean of tight duplicates from a representative experiment (n = 3) is shown (d, e). f IC50 values calculated from three independent experiments (see e for a representative experiment) were normalized to those of nonbiotinylated HCRT1–13 and shown as means ± SEMs.
Unlike the “empty” DQ6 on yeast, the soluble DQ6 protein with linked CLIP87–101 requires thrombin cleavage and DM catalysis for CLIP removal and peptide loading [5]. Despite the different conditions, the two empirical results showed a strong correlation (R2 = 0.6878, Fig. 4b), which was slightly higher than the correlation (R2 = 0.5896, Fig. 4c and R2 = 0.5024, Supplementary Fig. 4b) between the data acquired using the RIPPA approach and the prediction by NetMHCIIpan-4.0. Both empirical approaches showed >50%Competition for HCRT1–15, HCRT21–35, HCRT25–39, HCRT53–67, HCRT57–71, HCRT81–95, and HCRT85–99 peptides that carry previously determined DQ6-binding registers, whereas RIPPA further showed >75%Competition for HCRT89–103 (Fig. 4a, b), yielding 100% coverage of previously determined registers [4, 5, 44]. Titration of [HCRT89–103] in the competition assay suggested that >75%Competition can be obtained by yeast display when [competitor]:[indicator] = 10 (Fig. 4d), but using soluble proteins only when [competitor]:[indicator] = 100 (Fig. 4e and Supplementary Fig. 4c). Both empirical approaches confirmed HCRT89–103 as a DQ6 ligand, albeit with the relatively high IC50 suggested by the capture ELISA (Fig. 4d–f), consistent with the observation that the HCRT89–103 amino acid sequence includes the register NHAAGILTM [4, 5, 44]. Notably, although the indicator signal window in the flow cytometric measurements was relatively small (see Fig. 3a, b), the sensitivity of this approach for the identification of MHC-II ligands was as good as or occasionally even better than that of its orthogonal counterpart, ELISA (Fig. 4b).
Given the confidence inspired by both empirical binding datasets, we next evaluated the performance of the predicted binding results. The prediction algorithm that was trained mainly on eluted ligand (EL) mass spectrometry data performed slightly worse than the one that was trained mainly on binding affinity (BA) measurement data. HCRT1–15 was not ranked in the top 10%, a default cutoff used by NetMHCIIpan-4.0 to suggest binders, by either method (Fig. 4a), thus missing potential T cell epitopes using the register (LPSTKVSWA) previously proven to bind DQ6 by X-ray crystallography [45] and by both of our empirical assays (Fig. 4d–f). Moreover, HCRT21–35 was not ranked in the top 10% by either method. In addition, the EL-based algorithm predicted a ranking for HCRT89–103 below the cutoff and a ranking for HCRT113–131 above the cutoff, with the latter likely being a false positive.
Overall, the comparison validates RIPPA as an efficient and accurate method for the identification of DQ6 ligands derived from pre-pro-HCRT. Empirical data obtained from the RIPPA and ELISA methods reciprocally validate each other and demonstrate the advantages of these methods over prediction algorithms for the determination of MHC-II binders.
Identification of DR4 peptide ligands from the SARS-CoV-2 S protein
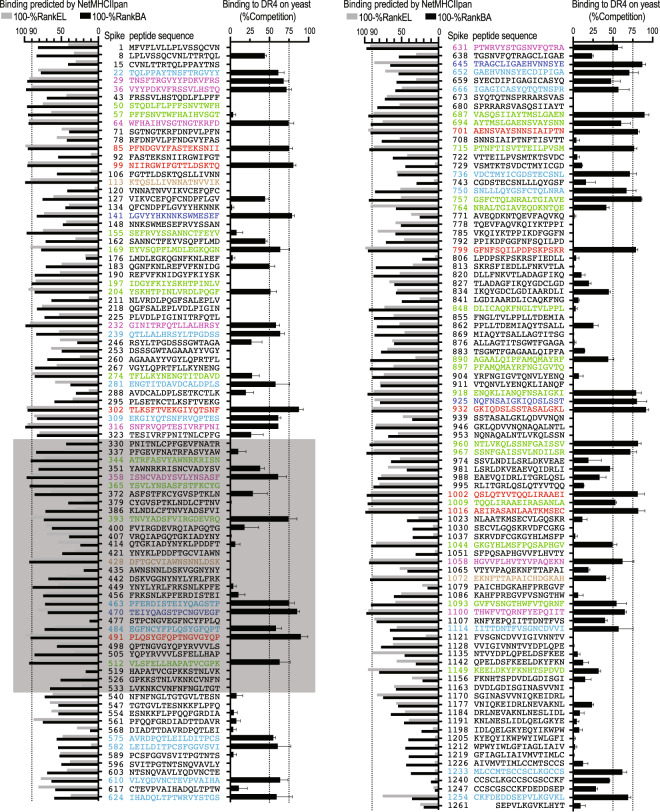
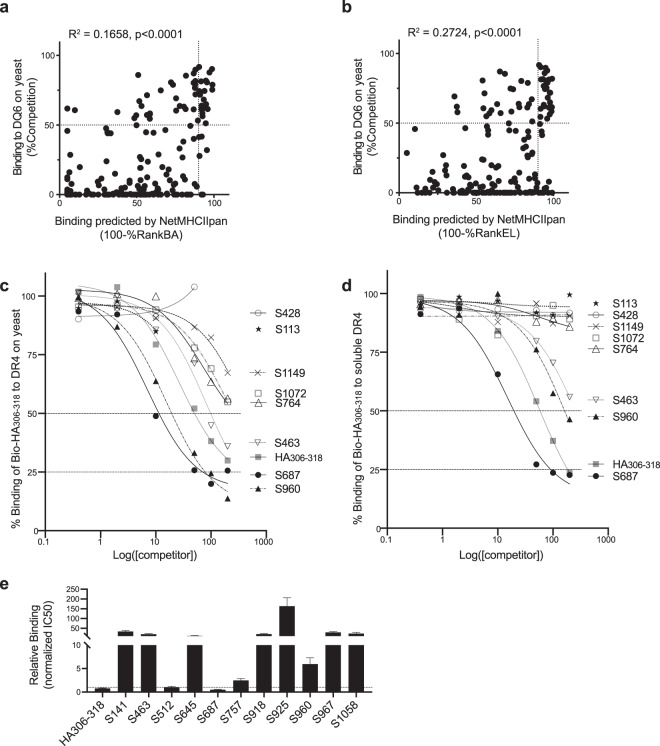
We next applied RIPPA to identify MHC-II ligands from the SARS-CoV-2 S protein. As a demonstration, we screened a set of 181 overlapping 17-mer peptides (Fig. 5) for DR4 ligands. DRB1*04:01 has a frequency of ~10% in the population of European descent and 1–2% in other populations (https://bioinformatics.bethematchclinical.org). The use of a 96-well polymerase chain reaction (PCR) plate enabled simultaneous culture of 96 wells of sufficient yeast cells expressing “empty” DR4, and allowed screening for up to 94 nonbiotinylated peptides as competitors (one per well) against the indicator peptide binding to the “empty” DR4 expressed on yeast. Eighteen peptides showed >75%Competition, and 34 showed 50–75%Competition by RIPPA (Fig. 5). A total of 31 of these 52 peptides (59.6%) were predicted to rank in the top 10% by at least one of the two NetMHCIIpan-4.0 algorithms (BA and EL). The insufficient coverage of RIPPA-identified DR4 ligands by prediction was also reflected by the lower correlation (RIPPA vs BA, R2 = 0.1658; RIPPA vs EL, R2 = 0.2724; Fig. 6a, b) than that observed in the pre-pro-HCRT analysis (RIPPA vs BA, R2 = 0.5896, Fig. 4c and RIPPA vs EL, R2 = 0.5024, Supplementary Fig. 4b). Notably, 16 peptides predicted to rank in the top 10% by at least one of the two algorithms showed <50%Competition by RIPPA (Fig. 5).
Fig. 5. DR4-binding peptides derived from the SARS-CoV-2 S protein identified by yeast display vs computational prediction.
Yeast cells expressing “empty” DR4 were incubated with 20 μM Bio-HA306–318 and the indicated nonbiotinylated spike peptide (200 μM) at pH 5.0 and 30 °C for 20 h, and were then analyzed (as in Fig. 3f). The error bars indicate the SEM of four independent experiments. Binding ranks of each spike peptide predicted by NetMHCIIpan-4.0 are shown to the left (as in Fig. 4a). The number in front of each peptide indicates the start position in the S precursor. Red: peptides that were ranked in the top 10% by both prediction algorithms and showed >75%Competition by RIPPA; magenta: peptides that were ranked in the top 10% by both prediction algorithms and showed 50–75%Competition by RIPPA; brown: peptides that were ranked in the top 10% by both prediction algorithms and showed <50%Competition by RIPPA; blue: peptides that were ranked below the cutoff (10%) by both prediction algorithms and yielded >75%Competition by RIPPA; cyan: peptides that were ranked below the cutoff by both prediction algorithms and yielded 50–75%Competition; green: peptides that were ranked in the top 10% by either prediction algorithm. The gray area indicates peptides derived from the receptor-binding domain (RBD) of the S protein.
Fig. 6. RIPPA performs better than selected computational prediction algorithms.
a, b Correlation analysis of binding data acquired using “empty” DR4 displayed on yeast vs NetMHCIIpan-4.0 predictions using the BA (b) or EL (c) algorithms. c Yeast cells were incubated with 20 μM Bio-HA306–318 and various concentrations of competitor peptides and were then analyzed (as in Fig. 4d). d Soluble DR4 (10 nM) was incubated with 20 μM Bio-HA306–318 and various concentrations of competitor peptides in the presence of 50 nM soluble DM. DR4-bound Bio-HA306–318 under each condition was quantified by capture ELISA using time-resolved fluorescence and was then analyzed (as in Fig. 4e). The mean of tight duplicates from a representative experiment (n = 3) is shown (c, d). e Soluble DR4 (10 nM) was incubated with 1 μM Bio-HA306–318 and various concentrations of competitor peptides in the presence of 50 nM soluble DM. DR4-bound Bio-HA306–318 under each condition and was then analyzed (see Supplementary Fig. 5a). The IC50 values of each competitor calculated from three independent experiments were normalized to those of nonbiotinylated HA306–318 and shown as means ± SEMs.
To estimate the false-positive or false-negative discovery rates (FDRs) based on the identifications from RIPPA and the BA or EL rank predictions, we quantified the %Competition of the aforementioned 68 peptides using soluble MHC-II in a capture ELISA (Supplementary Table 1). Similar to the DQ6-HCRT binding measurements (Fig. 4b, d, e), RIPPA and ELISA showed noticeable differences for the %Competition (Supplementary Table 1) and inhibition curves (see Fig. 6c, d for examples) representing DR4-Spike binding. However, for 16 DR4-binding candidates (excluding S1058–1074) that showed discrepancies between the RIPPA data and computational prediction data, the ELISA method validated the RIPPA method for the identification of all 10 DR4 binders (IC50 ranks in Figs. 6e) and five of the six nonbinders (Supplementary Fig. 5a). A likely RIPPA-identified false positive, S652–668, showed <50%Competition when [competitor]:[indicator] = 100 by ELISA. To eliminate experimental error caused by selection of certain peptides, we synthesized alternative peptides spanning the six nonbinder peptide regions and tested their competitive binding to soluble DR4 or “empty” DR4 expressed on yeast. All alternative peptides showed 50%Competition under the experimental conditions (Supplementary Fig. 5b), confirming that the corresponding 17-mers were false positives by at least one of the predictions. Assuming the capture ELISA also identified the remaining 113 (=181 − 68) peptides as nonbinders, RIPPA (FDR~4.97%, 9/181, 8 false positives) performed at least as well as ELISA (FDR ≥ 3.87%, 7/181, all false negatives), and both yielded more accurate identifications than the NetMHCIIpan-4.0 algorithms (BA: FDR~16.6%, 30/181, 17 false negatives; EL: FDR~15.5%, 28/181, 23 false negatives; see Supplementary Table 1). This result is reminiscent of the lower false-negative rate observed in the DQ6-HCRT binding data from the RIPPA method (0%) vs the capture ELISA method (3.33%, 1/30) or the BA (6.67%, 2/30) and EL (10%, 3/30) rank predictions (see Fig. 4). RIPPA is thus a better approach to rapidly identify the highest number of lead MHC-II binders for downstream analysis.
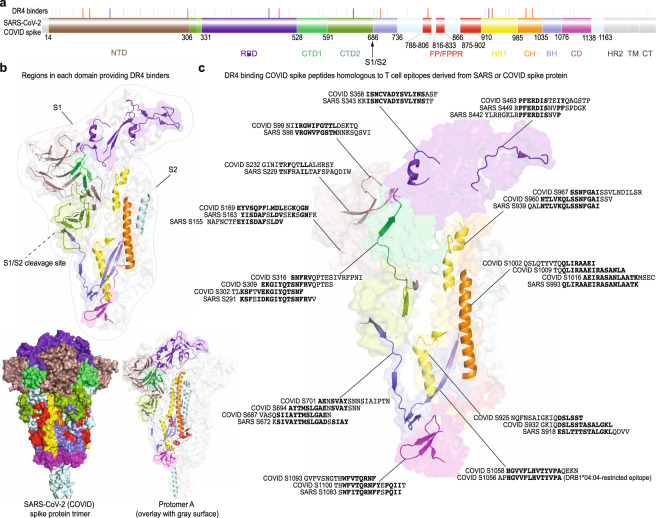
Considering all four datasets, we identified 45 lead DR4 binders (Supplementary Table 1), 27 from the S1 subunit and 18 from the S2 subunit (Figs. 5 and 7a, b). S1 contains N-terminal and C-terminal domains and a receptor-binding domain, which is essential for viral attachment and transmission [46]. The S2 subunit, containing fusion peptides, two heptad repeats (HR1 and HR2) and a central helix, functions to bring viral and cellular membranes into close proximity for fusion and infection [47]. DR4 binders span each of these domains (except for HR2) without particular location or structural preferences, suggesting a wide range of candidate immunogenic targets for vaccine design (Fig. 7a, b). Seventeen DR4-restricted SARS-CoV-1 S peptides have previously been suggested to contain putative T cell epitopes [13]; 13/17 (76.5%) of these SARS-CoV-1 S peptides share at least 4 homologous amino acids (aas) with 20 of the 45 SARS-CoV-2 S-derived DR4 binders (Fig. 7c). Another DR4 binder, S1058–1074 (with a BA ~20× weaker than that of HA306–318, Fig. 6e), shares 13 aa residues with a SARS-CoV-2 S epitope (Fig. 7c) that can stimulate CD4+ T cells isolated from an unexposed individual carrying a DR4 allelic variant [6]. These identifications provide the molecular basis for the speculation (based on recent studies [6–9]) that cross-reactive memory CD4+ T cells likely arose prior to the COVID-19 pandemic. This initial set of DR4-restricted epitope candidates thus enables peptide/DR4 tetramer synthesis for probing cross-reactive CD4+ T cell clones from DR4+ individuals.
Fig. 7. DR4 binders derived from the SARS-CoV-2 (COVID) S protein.
a Mapping DR4 binders to different domains of the S precursor. Each vertical bar indicates the exact starting position of a DR4-binding S peptide (the colors of the vertical bars match Fig. 5 and Supplementary Table 1). NTD N-terminal domain, RBD receptor-binding domain, CTD1/2 C-terminal domain 1/2 (or subdomain 1/2, SD1/2), FP/FPPR fusion peptide and fusion peptide proximal region [53], HR1/2 heptad repeat 1/2, CH central helix, BH b-hairpin [54], CD connector domain (or subdomain 3, SD3), TM transmembrane domain, CT cytoplasmic tail. b Regions on protomer A of the COVID S protein that contain DR4-binding peptides. The cartoon (over surface) presentation of each domain uses the same color scheme as in a. c Comparison of SARS S-derived DR4 binders with previously identified candidate DR4-restricted T cell epitopes [6, 13] that are from either the SARS-CoV-1 (SARS) or SARS-CoV-2 (COVID) S protein. Identical or conserved residues are bolded. The starting position of each COVID S or SARS S peptide and the cartoon (over surface) presentation of the corresponding COVID S peptide on protomer A are indicated. The structure [47] was obtained from PDB ID 6XR8 with an emphasis on protomer A. The individual showing positive CD4+ T cell responses to the SARS-CoV-2 S epitope carried DR4 allelic subtype DRB1*04:04 [6], as indicated in parentheses.
Discussion
The global COVID-19 health emergency has led to intensive efforts to understand CD4+ T cell-mediated immunity against SARS-CoV-2 [6–9] for strategic guidance of vaccine design and immunotherapeutic approaches. The use of computational algorithms that have been trained on existing peptide elution and binding data is the quickest way to predict MHC-II ligands derived from candidate SARS-CoV-2 antigens for use in T cell assays [48, 49]. Given the false-positive rate [17–20] and incomplete coverage of ligands from selected antigens in the prediction data, as observed here, it is still essential to experimentally determine MHC-II binding and verify HLA restriction of T cell epitopes prior to downstream experimental and clinical investigations. In this study, we developed a RIPPA method to quickly examine the binding of the spectrum of antigen-derived peptides to a given MHC-II allelic protein displayed by yeast cells. Unlike other experimental setups, this method does not require time-consuming steps, including the construction and preparation of cell lines expressing a single target MHC-II allele and the labor-intensive isolation and preparation of MHC-II proteins.
The engineering of MHC α and β polypeptides as a single-chain fusion to Aga2p in yeast was previously developed, with the aim of establishing a high-throughput surface display platform to study MHC–peptide–TCR interactions. However, under most conditions, point mutations in MHC alleles and covalently linked stabilizer peptides are necessary for appropriate protein folding [14–16, 38–41, 50]. To display MHC-II molecules in their native heterodimeric form while avoiding unnecessary genetic modifications, we adopted a bidirectional expression construct [33, 34] that enables α and β chains to be expressed separately in yeast. We also utilized a previously verified LZ dimerization motif [31, 32] to facilitate chain pairing. Our construct yielded “genetically empty” yet correctly folded DR or DQ α/β heterodimers anchored at the yeast cell surface to mimic their native counterparts for binding antigenic peptides. Both “empty” DR4 and DQ6 proteins were fully functional and accommodated biotinylated indicator peptides under various conditions, allowing experimentation under both the optimum time and pH conditions for MHC-II peptide binding and the habitual temperature for yeast survival. In addition, molecules such as DQ6, which typically rely on DM catalysis to remove a preloaded stabilizer peptide (e.g., CLIP), can spontaneously bind the biotinylated indicator peptide on yeast and be detected using flow cytometry. As described before [33, 41, 51] and demonstrated here, flow cytometric data can be mathematically converted to calculate binding parameters, i.e., kobs, Kd,app and IC50. In particular, the advantage of this endpoint measurement for determining the relative binding capacities of different peptides has been demonstrated. These features enable the study of competitive peptide binding at the yeast cell surface and the development of a scalable RIPPA approach for the identification of MHC-II ligands. Here, we validated the efficiency and accuracy of the RIPPA method and then identified DR4 ligands derived from the SARS-CoV-2 S protein as a model antigen.
We identified 45 DR4-binding peptides spanning most major domains of the SARS-CoV-2 S protein, one of which has substantial overlap with a recently identified CD4+ T cell epitope, though its HLA restriction is not empirically defined [6]. Notably, 44 of these DR4 binders were identified by RIPPA (>50%Competition), whereas 32 were predicted by NetMHCII-4.0 (ranked in the top 10% by both the BA and EL algorithms). The relatively lower coverage (71.1% vs 97.8%) in the computational prediction approach reemphasizes the importance of empirical examination of MHC-II ligands. Knowing the capacity of MHC-II molecules to bind peptides derived from multiple domains of a microbial antigen, as demonstrated here for DR4 ligands, provides useful information for vaccine design, particularly vaccines with subunit- or peptide-based agents. Viruses, including coronaviruses, mutate naturally for increased fitness [52]. Therefore, combinatorial inclusion of alternative epitopes or selection of candidates with conserved amino acids across mutants or across different strains of the same species of virus is likely optimal for vaccination.
The yeast display platform is also amenable to coupling with techniques, such as directed evolution [40], single-cell sequencing [10], and TCR signaling assays [11] for characterization of TCRs and T cell epitopes or mimotopes, given the ability of yeast to display both “empty” and peptide-linked constructs of noncovalent MHC-II α/β heterodimers. In addition, it remains possible that these native-like MHC-II molecules displayed on yeast can support antigen presentation to CD4+ T cells, as previously tested [38], for the potential development of “artificial” APCs for scientific and therapeutic purposes. Given the fast pace of technological and computational development and the rapidly evolving natural environment, RIPPA and its optimized versions are likely to provide significant advantages for future MHC–peptide–TCR research.
Methods
Materials
The plasmids Z47 and ptDR1 were gifts received from Dr. Eric Boder (University of Tennessee, Knoxville) [33, 34]. HotStarTaq DNA Polymerase was purchased from Qiagen (Valencia, CA). Vent® DNA Polymerase, restriction enzymes, DNA ligase, and DH5alpha competent E. coli were obtained from New England Biolabs (NEB, Beverly, MA). Oligonucleotides used as PCR primers were synthesized by Integrated DNA Technologies, Inc. (IDT, Coralville, IA). The DNA sequencing service was provided by MCLAB (South San Francisco, CA). Five biotinylated peptides, namely, Bio-HA306–318 (biotin-Ahx-PKYVKQNTLKLAT), Bio-MHC-Ia (biotin-Ahx-APWIEQEGPEYWDQE) [28], HCRT1–13-Bio (MNLPSTKVSWAAVK-Ahx-biotin), MHC-Ia-Bio (APWIEQEGPEYWDQEK-Ahx-biotin), and Bio-EBV486–500 (biotin-GGG-RALLARSHVERTTDE), and the nonbiotinylated HA306–318 and HCRT1–13 peptides, were synthesized by GenScript (Piscataway, NJ). The 30 15-mer overlapping peptides (offset by 4 aa, Fig. 4a) derived from pre-pro-HCRT were synthesized by GenScript. The 181 overlapping 17-mer peptides (the last one contained 13 aa, offset by 7 aa; Fig. 5) derived from the SARS-CoV-2 S protein were ordered from BEI Resources (https://www.beiresources.org). Other tested spike peptides (Supplementary Fig. 5) were synthesized by Apeptide Co., Ltd. (Shanghai, China). Peptides were dissolved in DMSO at concentrations of >5 mM, stored in a −20 °C freezer and diluted in sterile water prior to use. The mAbs mouse anti-DRαβ (clone L243) and mouse anti-DQαβ (clone SPV-L3) were affinity purified from ascites, as described previously [33, 34]. The rabbit anti-HA-tag mAb was purchased from Sigma (St. Louis, MO). The mouse anti-c-Myc-Tag mAb was purchased from Cell Signaling. Alexa Fluor 647-conjugated streptavidin was purchased from Invitrogen. Highly cross-adsorbed secondary antibodies, including Alexa Fluor 488 goat anti-rabbit IgG (H + L) and Alexa Fluor 647 goat anti-mouse IgG (H + L), were purchased from Thermo Fisher Scientific (Waltham, MA). Other chemical reagents were purchased from Thermo Fisher Scientific unless otherwise indicated.
Preparation of yeast display constructs
The nucleotides encoding the HA306–318 peptide in Z47 were first removed to construct a plasmid that allowed the expression of “empty” DR4 on yeast (plasmid synthesis was performed by GenScript). To construct the “empty” DR4-LZ and HA306–318/DR4-LZ plasmids, the Fos and Jun LZ dimerization motifs used previously [5, 24, 39] were fused to the C termini of the DR4β and DR4α chains, respectively. The c-Myc epitope tag was fused to the C terminus of the Jun motif (plasmid synthesis was performed by GenScript). The backbone yeast shuttle vector used for surface expression of “empty” DQ6-LZ or peptide/DQ6-LZ was based on the plasmid ptDR1 constructed previously [33]. A PCR fragment containing the extracellular domain of HLA-DQA1*0102 with a C-terminal Fos motif was cloned into ptDR1 in place of the original expression cassette coding for the DR1 β chain via the XmaI and SpeI restriction sites. This created an in-frame fusion of DQA1 to the N terminus of Aga2p (pDQ6α). A second PCR fragment containing the extracellular domain of HLA-DQB1*0602 with a C-terminal Jun motif was cloned into pDQ6α in place of the original expression cassette coding for the DR1 α chain via the EagI and SalI restriction sites to construct the ptDQ6-LZ plasmid that directed the expression of “empty” DQ6-LZ on yeast. Plasmids directing the expression of peptide/DQ6-LZ constructs, including CLIP87–101/DQ6 (CLIP87–101 aa sequence: PVSKMRMATPLLMQA), HA273–286/DQ6 (HA273–286 aa sequence: RALLARSHVERTTD), and HCRT87–97/DQ6 (HCRT87–97 aa sequence: SGNHAAGILTM), were synthesized (the CLIP87–101/DQ6-LZ and HA273–286/DQ6-LZ constructs were synthesized by GenScript) using a similar strategy, with the peptide sequence located upstream of the DQ6 β chain gene.
Protein expression in yeast
Plasmids carrying the tryptophan nutrition marker gene (TRP+) were then transformed into the yeast parent strain EBY100 (URA+ and TRP−) by electroporation following the BioRad MicroPulser protocol. After 2 days at 30 °C, single yeast colonies can grow on agar plates containing tryptophan dropout medium, e.g., SD-CAA (2% (w/v) glucose, 0.67% (w/v) yeast nitrogen base without amino acids, 0.062% (w/v) Ura/Trp dropout casamino acids, 38 mM Na2HPO4, and 62 mM NaH2PO4 (pH 6.0)). Two milliliters of SD-CAA minimal medium was then inoculated with a single yeast colony and cultured overnight at 30 °C with shaking at 225 r.p.m. to an OD600 of 2.5–5.0. To induce GAL1–10-driven protein expression in yeast, 107 cells were harvested and switched to 2 ml SG-CAA medium (in which glucose was replaced with galactose). After 18 h of induction at 30 °C, a sufficient number of yeast cells per sample were collected by centrifugation at 2500 × g for 3 min, and were then washed and prepared for analysis of protein expression or peptide binding.
Immunofluorescence staining and flow cytometry
The expression of “empty” or peptide-linked MHC-II was assessed using immunofluorescence labeling and flow cytometry. Briefly, galactose-induced yeast cells were first costained with primary mAbs, including mouse mAbs L243 (for DR4) or SPV-L3 (for DQ6) and a rabbit anti-HA-tag mAb (~10 μg/ml for each mAb), at room temperature for 30 min and then on ice for 30 min. Cells were then washed with 300 μl of ice-cold phosphate-buffered saline (PBS) + 1% (w/v) bovine serum albumin (BSA), and double labeled with highly cross-adsorbed secondary antibodies (1:100 dilution; Alexa Fluor 488 goat anti-rabbit IgG (H + L) and Alexa Fluor 647 goat anti-mouse IgG (H + L)) on ice for 1 h. To further validate that both chains of MHC-II were expressed by yeast, a mouse anti-c-Myc-tag mAb (1:500 dilution) and a rabbit anti-HA-tag mAb were used in the primary labeling step. After labeling, yeast cells were analyzed in a FACSCalibur flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ) to detect fluorescence signals corresponding to the expression of MHC-II proteins or epitope tags. At least 100,000 cell events gated by forward and side scatter were collected per sample. Flow cytometric data were analyzed using FlowJo software (version 10.6.0, BD).
Loading exogenous peptides into “empty” MHC-II molecules displayed on yeast
A total of 6 × 105 galactose-induced yeast cells expressing “empty” MHC-II molecules were collected by centrifugation at 2500 × g for 3 min and resuspended in 40 μl of the following solutions: citrate buffer (40 mM citric acid and sodium citrate (pH 5.0), 150 mM NaCl, and 1% (w/v) BSA) and PBS (pH 7.4; 137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4, and 1% (w/v) BSA). Biotinylated peptides, namely, Bio-HA306–318 and Bio-MHC-Ia for DR4 or HCRT1–13-Bio and MHC-Ia-Bio for DQ6, were then added to the solution at 20 μM prior to incubation under the desired conditions. To determine the kinetics of peptide binding, 6 × 105 galactose-induced yeast cells were collected and resuspended in 40 μl of 40 mM citrate buffer (pH 5.0), incubated with biotinylated peptides for different durations, and collected by centrifugation at 2500 × g for 3 min for analysis by flow cytometry. To determine the apparent BA of MHC-II peptide binding at the yeast cell surface, yeast cells were incubated with 0, 0.2, 0.5, 2, 5, 20, 50, and 100 μM Bio-HA306–318 and Bio-MHC-Ia (for DR4) or HCRT1–13-Bio and MHC-Ia-Bio (for DQ6) in 40 mM citrate buffer (pH 5.0) at 30 °C for 20 h. The unrelated biotinylated peptides MHC-Ia-Bio and Bio-MHC-Ia were used as negative controls. The yeast parent strain EBY100 was used to evaluate the BG staining of the biotinylated indicator. The reaction tubes were sealed with parafilm before incubation to prevent variations in the culture volume that may affect the final peptide concentration in the time course or concentration titration studies. After incubation, yeast cells were washed twice with 300 μl of ice-cold PBS + 1% BSA before staining with streptavidin-AF647 diluted 1:200 in 50 μl of PBS + 1% BSA on ice for 1 h. Cells were then washed with 300 μl of ice-cold PBS + 1% BSA twice and were finally resuspended in 300 μl of ice-cold PBS + 1% BSA for analysis in a BD FACSCalibur flow cytometer (BD Biosciences). For simultaneous detection of both cell surface MHC-II proteins and biotinylated peptides, cells with bound biotinylated peptides were first stained with a rabbit anti-HA-tag mAb on ice for 30 min and were then double labeled with highly cross-adsorbed Alexa Fluor 488 goat anti-rabbit IgG (H + L) and streptavidin-AF647 in PBS + 1% BSA on ice for 1 h. Flow cytometric data was analyzed using FlowJo.
Evaluation of binding kinetics and equilibrium in yeast
Like other endpoint approaches, the flow cytometry-based yeast display approach is suboptimal for measuring kinetic parameters. However, if the experiment is designed appropriately (e.g., [pep] ≫ [MHC]), peptide exchange on the surface of yeast cells can be approximated as a pseudo-first-order equimolar reaction, as described previously [24]: [pepMHC] = [pepMHC]eq × (1 − exp(−kobst)) and [pepMHC]eq = [pep] × [pepMHC]max/(Kd + [pep]). The observed rate constant (kobs) and the apparent equilibrium dissociation constant (Kd,app) can be calculated by fitting multiple time point data and equilibrium binding data, respectively. In a 40 μl reaction containing 6 × 105 galactose-induced yeast cells, the approximate [MHC] = (6 × 105 cells × 105 MHC molecules/cell)/(6.02 × 1023 MHC molecules/mol)/40 µl = 2.5 nM, given an average of ~105 copies of recombinant protein Aga2p per yeast cell [29]. The [pep] ≫ [MHC] assumption was confirmed to be valid in the 0.2–100 μM range of indicator peptides. To calculate kobs, peptide–MHC binding was quantified as the background-subtracted median fluorescence intensity (MFI) of the streptavidin staining signal (i.e., [pepMHC] = MFISA,indicator − BG) and was plotted against the incubation time. Data were fitted to the equation [pepMHC] = [pepMHC]eq × (1 − exp(−kobst)). To calculate Kd,app, incubation was continued for 20 h to allow equilibrium to be attained, and [pepMHC]eq at each peptide concentration was plotted against [pep]. Data were fitted to the equation [pepMHC]eq = [pep][pepMHC]max/(Kd + [pep]). Fitting was performed using GraphPad Prism.
Peptide competition assay using yeast displaying MHC-II molecules
A total of 6 × 105 galactose-induced yeast cells were incubated at pH 5.0 and 30 °C for 20 h with 20 μM biotinylated indicator peptides in the presence of various concentrations of a competitor peptide to determine an appropriate competitor concentration for the competition assay. The nonbiotinylated competitor peptides used for DR4 and DQ6 were HA306–318 and HCRT1–13, respectively. After incubation, yeast cells were washed with PBS + 1% BSA, stained with streptavidin-AF647 and analyzed by flow cytometry, as described above. The %Binding in the presence of competitors was quantified as [(MFIwith competitor − BG)/(MFIwithout competitor − BG)] × 100% and plotted against the competitor concentration to determine the IC50. Data were fitted using nonlinear regression with the one site-fit logIC50 equation in GraphPad Prism. The peptide competition assay by yeast display revealed that a [competitor]:[indicator] ratio >2 yielded quite a robust competition signal, and we chose the highest amenable ratio ([competitor]:[indicator] = 10) for RIPPA experiments. At this ratio, %Competition was calculated as 100% − %Binding to reflect binding of the competitor to MHC-II molecules displayed on yeast. Yeast is a living system. Changes in yeast cell states (viability, age, morphology, protein induction level, etc.) are the main contributors to experimental uncertainty and variations. Therefore, to obtain reliable binding values, we fitted replicated data points within the same experiments that used the same batch of yeast cells. To determine relative binding capacities (%Competition values), we performed independent experiments and maintained a consistent yeast growth and induction routine with minimal technical changes. Another factor that may cause experimental uncertainty by influencing yeast cell states is the peptide components. It is highly likely that some peptides or unidentified contents in the peptide preparation are toxic to yeast cells and can cause cell death, thereby increasing the autofluorescence signal in the flow cytometric data. This possibility was avoided by strategic gating on the main population in a dot plot (see Fig. 3b for an example).
Identification of DQ6-binding peptides in HCRT
Sufficient amounts of galactose-induced yeast cells displaying “empty” DQ6 were collected by centrifugation at 2500 × g for 3 min and resuspended in 40 mM citrate buffer (pH 5.0) at a density of 1.5 × 104 cells/μl. Then, 20 μM HCRT1–13-Bio peptide was added to make a master mix of the reaction solution. Forty-microliter aliquots were aspirated from the solution, and each aliquot was supplemented with a 15-mer peptide (each at 200 μM) derived from pre-pro-HCRT. In parallel, cultures of yeast without biotinylated peptides and cultures of yeast with MHC-Ia-Bio were also prepared and used as the BG and negative controls, respectively. The reaction was carried out under acidic conditions at 30 °C for 20 h. DQ6-associated HCRT1–13-Bio was labeled with streptavidin-AF647 and analyzed by flow cytometry as described above. The binding of a competitor peptide to “empty” DQ6 displayed on yeast was quantified as follows: %Competition = 100% − [(MFIwith competitor − BG)/(MFIwithout competitor − BG)] × 100%.
High-throughput identification of DR4-binding peptides in the SARS-CoV-2 spike protein
Here, the competitive binding assay was scaled to a 96-well format. Sufficient amounts of galactose-induced yeast cells displaying “empty” DR4 were mixed with 20 μM Bio-HA306–318 in 40 mM citrate buffer (pH 5.0) at a density of 1.5 × 104 cells/μl. Forty microliters of the master mix was dispensed into each well of a 96-well PCR plate (Eppendorf twin.tec® PCR Plate 96), except for the negative control well and the well with only galactose-induced yeast cells, using a multichannel pipette. Up to 94 synthesized nonbiotinylated peptides derived from the SARS-CoV-2 S protein were then added into the wells of the 96-well plate to a final concentration of 200 μM (one peptide per well). The reaction plate was sealed with plate sealer (Eppendorf Storage Foil) to prevent variations in the culture volumes that may affect the peptide concentrations, and the plate was then incubated at 30 °C for 20 h. After incubation, the yeast cells in the 96-well PCR plate were washed three times with 150 μl of ice-cold PBS + 1% BSA using a multichannel pipette before staining with streptavidin-AF647 diluted 1:200 in 50 μl of PBS + 1% BSA on ice for 1 h. Cells were then washed with 150 μl of ice-cold PBS + 1% BSA three times and were finally resuspended in 300 μl of PBS + 1% BSA for flow cytometric analysis. The binding of a competitor peptide to “empty” DR4 displayed on yeast was quantified as follows: %Competition = 100% − [(MFIwith competitor − BG)/(MFIwithout competitor − BG)] × 100%.
Purification of soluble recombinant proteins
Stable S2 insect cell lines secreting CLIP87–101/sDQ6, sDR4 or sDM recombinant proteins were previously constructed [24]. sDQ6 with covalently linked CLIP87–101 was cleaved by thrombin before the peptide competition experiment. sDR4 was engineered without any covalent peptides [24]. To purify proteins from S2 cell culture supernatant, S2 cells were first cultured in complete Schneider Drosophila medium (supplemented with 10% FBS and 2 mM glutamine, Thermo Fisher Scientific) and induced using 1 mM copper sulfate. The supernatant was then filtered through a 0.22 µm membrane to remove cell debris, and proteins were purified by affinity chromatography followed by size exclusion chromatography on a Superdex 200 Increase gel filtration column (GE Healthcare, Chicago, IL).
Peptide competition assay by capture ELISA
Soluble recombinant MHC-II proteins were incubated with a biotinylated indicator peptide with or without competitor peptides in the presence of sDM at 37 °C for 20 h. The pH 4.7 reaction buffer contained 100 mM acetate buffer (acetic acid and sodium acetate), 150 mM NaCl, 1% (w/v) BSA, 0.5% (v/v) IGEPAL CA-630 (Sigma), and 0.1% (w/v) NaN3. After incubation, the peptide exchange reaction was stopped by the addition of two volumes of neutralization buffer containing 100 mM Tris-Cl (pH 8.5), 150 mM NaCl, 1% (w/v) BSA, 0.5% (v/v) IGEPAL CA-630, and 0.1% (w/v) NaN3. The mixture was transferred to a precoated ELISA plate for capture of biotinylated peptide-loaded MHC-II by anti-MHC-II antibodies (L243 or SPV-L3). A DELFIA Eu-N1 Streptavidin System (PerkinElmer) was used to quantify the time-resolved fluorescence, as previously described [5, 24]. Peptide–MHC binding was quantified as background-subtracted europium fluorescence: [pepMHC] = Eu-SAindicator − BG. To calculate %Binding and %Competition and fit data the same equations described above and elsewhere were used [24].
Comparison between empirical data and NetMHCIIpan-predicted data
The NetMHCIIpan-4.0 server (http://www.cbs.dtu.dk/services/NetMHCIIpan) was used to predict the binding of HCRT or SARS-CoV-2 S peptides to MHC-II proteins. The NetMHCIIpan-4.0 server provides two separate ranking algorithms, EL and BA; the former algorithm is trained mainly on EL mass spectrometry data, and the latter is trained mainly on BA measurement data. EL ranks may consider BA data, although the weighting on BA data is quite small due to the massive amount of EL data generated to date. We compared both computational prediction results with the results of the two empirical methods. Correlations between RIPPA-generated data and prediction data were analyzed by plotting one set against the other on an XY plot, and correlations (R-squared values) and statistical significance (p values) were determined using correlation analysis in GraphPad Prism. A higher correlation (represented by a higher R-squared value) indicates a higher chance of the same peptide ranking higher, e.g., of being an MHC-II ligand, and vice versa, as determined by both datasets. Cutoff values that separate binders from nonbinders were arbitrarily selected as 90%Rank (default by NetMHCIIpan-4.0) in both computational prediction datasets and 50%Competition in both empirical datasets.
Supplementary information
Acknowledgements
We thank Dr. Eric T. Boder in the Department of Chemical and Biomolecular Engineering, University of Tennessee, Knoxville, for providing necessary plasmids; Cynthia Jia for assistance with the preparation of reagents; and Dr. Hong Jin, Dr. Robbert Van Der Most, and Dr. Sofia Buonocore for critical review of the manuscript. This work was funded by the Lucile Packard Foundation for Children’s Health and a research grant from GlaxoSmithKline. R.L. was funded by the China Scholarship Council Funding (CSC) during his academic research at the Stanford University School of Medicine.
Author contributions
R.L., W.J., and E.D.M. conceived the project, designed the experiments, analyzed the results, and wrote the manuscript. R.L. performed plasmid construction, yeast transformation, flow cytometry, and peptide binding on yeast with assistance from W.J. All authors agreed with the submission.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Rongzeng Liu, Wei Jiang
Contributor Information
Wei Jiang, Email: wjiang6@stanford.edu.
Elizabeth D. Mellins, Email: mellins@stanford.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00717-5.
References
- 1.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 2.La Gruta NL, Gras S, Daley SR, Thomas PG, Rossjohn J. Understanding the drivers of MHC restriction of T cell receptors. Nat Rev Immunol. 2018;18:467–78. doi: 10.1038/s41577-018-0007-5. [DOI] [PubMed] [Google Scholar]
- 3.Latorre D, Kallweit U, Armentani E, Foglierini M, Mele F, Cassotta A, et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature. 2018;562:63–8. doi: 10.1038/s41586-018-0540-1. [DOI] [PubMed] [Google Scholar]
- 4.Luo G, Ambati A, Lin L, Bonvalet M, Partinen M, Ji X, et al. Autoimmunity to hypocretin and molecular mimicry to flu in type 1 narcolepsy. Proc Natl Acad Sci USA. 2018;115:E12323–E12332. doi: 10.1073/pnas.1818150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang W, Birtley JR, Hung SC, Wang W, Chiou SH, Macaubas C, et al. In vivo clonal expansion and phenotypes of hypocretin-specific CD4(+) T cells in narcolepsy patients and controls. Nat Commun. 2019;10:5247. doi: 10.1038/s41467-019-13234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–501 e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Bert N, Tan AT, Kunasegaran K, Tham C, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–62. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 9.Braun J, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–4. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 10.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–92. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Wang C, Rubelt F, Scriba TJ, Davis MM. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020;38:1194–202. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newell EW, Davis MM. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat Biotechnol. 2014;32:149–57. doi: 10.1038/nbt.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, James E, Roti M, Huston L, Gebe JA, Kwok WW. Searching immunodominant epitopes prior to epidemic: HLA class II-restricted SARS-CoV spike protein epitopes in unexposed individuals. Int Immunol. 2009;21:63–71. doi: 10.1093/intimm/dxn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saligrama N, Zhao F, Sikora MJ, Serratelli WS, Fernandes RA, Louis DM, et al. Opposing T cell responses in experimental autoimmune encephalomyelitis. Nature. 2019;572:481–7. doi: 10.1038/s41586-019-1467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell. 2018;174:672–87 e627. doi: 10.1016/j.cell.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappazzo CG, Huisman BD, Birnbaum ME. Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction. Nat Commun. 2020;11:4414. doi: 10.1038/s41467-020-18204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Khodadoust MS, Olsson N, Wagar LE, Fast E, Liu CL, et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat Biotechnol. 2019;37:1332–43. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen M, Andreatta M, Peters B, Buus S. Immunoinformatics: predicting peptide–MHC binding. Annu Rev Biomed Data Sci. 2020;3:191–215. doi: 10.1146/annurev-biodatasci-021920-100259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020;19:2304–15. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 20.Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, et al. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity. 2019;51:766–79 e717. doi: 10.1016/j.immuni.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Nanaware PP, Jurewicz MM, Leszyk J, Shaffer SA, Stern LJ. HLA-DO modulates the diversity of the MHC-II self-peptidome. Mol Cell Proteomics. 2019;18:490–503. doi: 10.1074/mcp.RA118.000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodadoust MS, Olsson N, Wagar LE, Haabeth OA, Chen B, Swaminathan K, et al. Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature. 2017;543:723–7. doi: 10.1038/nature21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell AW, Ramarathinam SH, Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. 2019;14:1687–707. doi: 10.1038/s41596-019-0133-y. [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Strohman MJ, Somasundaram S, Ayyangar S, Hou T, Wang N, et al. pH-susceptibility of HLA-DO tunes DO/DM ratios to regulate HLA-DM catalytic activity. Sci Rep. 2015;5:17333. doi: 10.1038/srep17333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. 2010;185:4189–98. doi: 10.4049/jimmunol.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterbye T, Nielsen M, Dudek NL, Ramarathinam SH, Purcell AW, Schafer-Nielsen C, et al. HLA class II specificity assessed by high-density peptide microarray interactions. J Immunol. 2020;205:290–9. doi: 10.4049/jimmunol.2000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Justesen S, Harndahl M, Lamberth K, Nielsen LL, Buus S. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 2009;5:2. doi: 10.1186/1745-7580-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung SC, Hou T, Jiang W, Wang N, Qiao SW, Chow IT, et al. Epitope selection for HLA-DQ2 presentation: implications for celiac disease and viral defense. J Immunol. 2019;202:2558–69. doi: 10.4049/jimmunol.1801454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–7. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 30.Boder ET, Jiang W. Engineering antibodies for cancer therapy. Annu Rev Chem Biomol. Eng. 2011;2:53–75. doi: 10.1146/annurev-chembioeng-061010-114142. [DOI] [PubMed] [Google Scholar]
- 31.Busch R, Pashine A, Garcia KC, Mellins ED. Stabilization of soluble, low-affinity HLA-DM/HLA-DR1 complexes by leucine zippers. J Immunol Methods. 2002;263:111–21. doi: 10.1016/S0022-1759(02)00034-0. [DOI] [PubMed] [Google Scholar]
- 32.Serra P, Garabatos N, Singha S, Fandos C, Garnica J, Solé P, et al. Increased yields and biological potency of knob-into-hole-based soluble MHC class II molecules. Nat Commun. 2019;10:4917. doi: 10.1038/s41467-019-12902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang W, Boder ET. High-throughput engineering and analysis of peptide binding to class II MHC. Proc Natl Acad Sci USA. 2010;107:13258–63. doi: 10.1073/pnas.1006344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boder ET, Bill JR, Nields AW, Marrack PC, Kappler JW. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol Bioeng. 2005;92:485–91. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 35.Adler LN, Jiang W, Bhamidipati K, Millican M, Macaubas C, Hung SC, et al. The other function: class II-restricted antigen presentation by B cells. Front Immunol. 2017;8:319. doi: 10.3389/fimmu.2017.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellins ED, Stern LJ. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr Opin Immunol. 2014;26:115–22. doi: 10.1016/j.coi.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinderknecht CH, Belmares MP, Catanzarite TL, Bankovich AJ, Holmes TH, Garcia KC, et al. Posttranslational regulation of I-Ed by affinity for CLIP. J Immunol. 2007;179:5907–15. doi: 10.4049/jimmunol.179.9.5907. [DOI] [PubMed] [Google Scholar]
- 38.Wen F, Esteban O, Zhao HM. Rapid identification of CD4+T-cell epitopes using yeast displaying pathogen-derived peptide library. J Immunol Methods. 2008;336:37–44. doi: 10.1016/j.jim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Wen F, Sethi DK, Wucherpfennig KW, Zhao H. Cell surface display of functional human MHC class II proteins: yeast display versus insect cell display. Protein Eng Des Sel. 2011;24:701–9. doi: 10.1093/protein/gzr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–87. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esteban O, Zhao H. Directed evolution of soluble single-chain human class II MHC molecules. J Mol Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 42.Han F, Lin L, Warby SC, Faraco J, Li J, Dong SX, et al. Narcolepsy onset is seasonal and increased following the 2009 H1N1 pandemic in China. Ann Neurol. 2011;70:410–7. doi: 10.1002/ana.22587. [DOI] [PubMed] [Google Scholar]
- 43.Partinen M, Saarenpää-Heikkilä O, Ilveskoski I, Hublin C, Linna M, Olsén P, et al. Increased incidence and clinical picture of childhood narcolepsy following the 2009 H1N1 pandemic vaccination campaign in Finland. PLoS ONE. 2012;7:e33723. doi: 10.1371/journal.pone.0033723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schinkelshoek MS, Fronczek R, Kooy-Winkelaar E, Petersen J, Reid HH, van der Heide A, et al. H1N1 hemagglutinin-specific HLA-DQ6-restricted CD4+ T cells can be readily detected in narcolepsy type 1 patients and healthy controls. J Neuroimmunol. 2019;332:167–75. doi: 10.1016/j.jneuroim.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Siebold C, Hansen BE, Wyer JR, Harlos K, Esnouf RE, Svejgaard A, et al. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc Natl Acad Sci USA. 2004;101:1999–2004. doi: 10.1073/pnas.0308458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–3. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM JR, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–92. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fast, E, Chen, B. Potential T-cell and B-cell epitopes of 2019-nCoV. bioRxiv: 10.1101/2020.02.19.955484 [Preprint]. (2020) [cited 20 Feb 2020]. Available from 10.1101/2020.02.19.955484.
- 49.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–.e672. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Starwalt SE, Masteller EL, Bluestone JA, Kranz DM. Directed evolution of a single-chain class II MHC product by yeast display. Protein Eng. 2003;16:147–56. doi: 10.1093/proeng/gzg018. [DOI] [PubMed] [Google Scholar]
- 51.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–70. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 52.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–.e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pal D. Spike protein fusion loop controls SARS-CoV-2 fusogenicity and infectivity. J Struct Biol. 2021;213:107713. doi: 10.1016/j.jsb.2021.107713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lokman SM, Rasheduzzaman M, Salauddin A, Barua R, Tanzina AY, Rumi MH, et al. Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: a computational biology approach. Infect. Genet. Evol. 2020;84:104389. doi: 10.1016/j.meegid.2020.104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.