Abstract
The cell-surface glycocalyx serves as a physiological barrier regulating cellular accessibility to macromolecules and other cells. Conventional glycocalyx characterization has largely been morphological rather than functional. Here, we demonstrated direct glycocalyx anchoring of DNA origami nanotiles and performed a comprehensive comparison with traditional origami targeting to the phospholipid bilayer (PLB) using cholesterol. While DNA nanotiles effectively accessed single-stranded DNA initiators anchored on the glycocalyx, their accessibility to the underlying PLB was only permitted by extended nanotile-to-initiator spacing or by enzymatic glycocalyx degradation using trypsin or pathogenic neuraminidase. Thus, the DNA nanotiles, being expelled by the physiologic glycocalyx, provide an effective functional measure of the glycocalyx barrier integrity and faithfully predict cell-to-cell accessibility during DNA-guided multicellular assembly. Lastly, the glycocalyx-anchoring mechanism enabled enhanced cell-surface stability and cellular uptake of nanotiles compared to PLB anchoring. This research lays the foundation for future development of DNA nanodevices to access the cell surface.
Keywords: DNA origami, glycocalyx, nanotechnology, metabolic labeling, cholesterol
Introduction and Background
The glycocalyx is a layer of plasma-membrane-associated biopolymers composed of proteoglycans, glycosaminoglycans, other glycoproteins, and glycolipids and can extend hundreds of nanometers from the external surface of the phospholipid bilayer (PLB).1−4 Through mechanisms, such as steric hindrance and electrostatic repulsion, the glycocalyx serves as a molecular barrier that effectively limits macromolecules, particles, and other cells from directly accessing the PLB.1,3,4 For example, the endothelial glycocalyx is an essential determinant of vascular permeability by excluding blood cells (such as erythrocytes and leukocytes) and plasma macromolecules from accessing the underlying endothelium.5−9 Similarly, the apical glycocalyx of the lung and intestinal epithelium acts as a selective barrier interfacing with the external environment and modulates epithelial interaction with foreign particles and microbes.10−12
The glycocalyx is a dynamic structure undergoing constant remodeling in response to environmental stimuli.13,14 Aberrant degradation of the endothelial glycocalyx leads to compromised vascular barrier function and contributes to vascular pathogenesis, such as atherosclerosis,15,16 stroke,17,18 hypertension,19,20 sepsis,7,21 and ischemia reperfusion injury.22,23 Moreover, shedding of the epithelial glycocalyx is associated with respiratory and intestinal infection, injury, and inflammation.10,11,24,25 Thus, characterization of the glycocalyx barrier integrity is of pivotal importance.
The glycocalyx is composed of delicate polymer structures at the nanometer scale. Accordingly, transmission electron microscopy (TEM) has long served as the gold standard for glycocalyx characterization.26−29 Alternative glycocalyx visualization uses fluorescent labeling or staining of particular glycocalyx components, albeit at a lower resolution compared to TEM.30,31 However, most of these approaches focus on evaluating the glycocalyx morphology rather than its function. Methods for specific assessment of the glycocalyx barrier function are limited and generally require sophisticated imaging techniques, such as intravital microscopy.1,7,32 Thus, there is a critical need for technologies to expedite direct examination of the glycocalyx barrier in living samples with high sensitivity and reproducibility.
Here, we explored the possibility of using DNA origami nanostructures as a functional measure of cell-surface glycocalyx barrier integrity. The DNA origami forms 2D and 3D nanostructures from the self-assembly of approximately 200 short single-stranded DNA (ssDNA), referred to as “staple strands”, based on a large ssDNA scaffold. Formation of the DNA origami allows highly predictable and reproducible assembly of biocompatible structures at the nanoscale and offers convenience for the incorporation of probe labeling (fluorescence and non-fluorescence) and sequence-selective targeting.33−37 With the capacity to carry a range of functionalities, DNA origami becomes a desirable candidate for assessing glycocalyx structures with thicknesses up to a few hundred nanometers.38
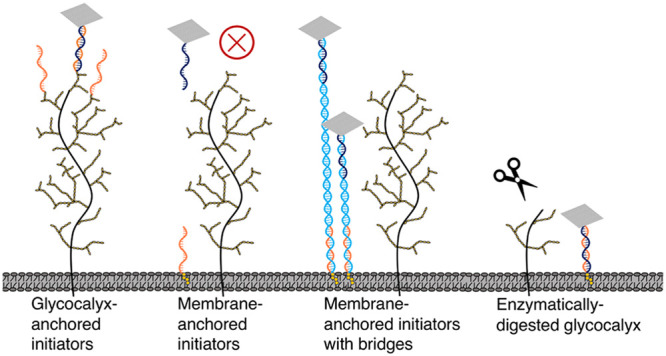
In this study, we assembled 2D DNA origami rectangles or “nanotiles”34 and examined ssDNA-initiator-mediated targeting of origami nanotiles to the cell surface. We found that nanotiles can only reach ssDNA initiators anchored on the glycocalyx but not those inserted directly on the PLB. The exclusion of nanotiles from the PLB by the physiologic glycocalyx was rescued either by using a technique reported previously by extending the spacing between the nanotile and PLB using a DNA duplex bridge33 or by enzymatic degradation of glycocalyx proteins via trypsin or neuraminidase, which is involved in type 2 diabetes,39 atrial stiffening,40,41 and viral infections.42,43 Our results establish DNA nanotiles as a functional measure of the glycocalyx barrier integrity.
Results and Discussion
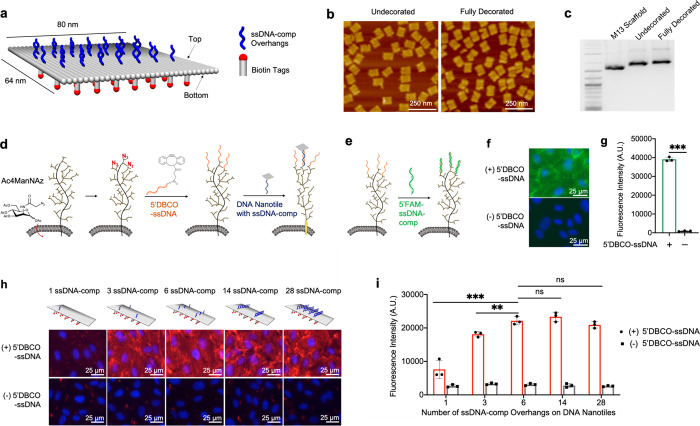
Flattened DNA origami nanotiles of 80 nm × 64 nm were prepared using a single-step annealing protocol using the bacteriophage M13mp18 ssDNA scaffold (Figure 1a, Supplementary Figure 1). The top surface of the nanotile was decorated with up to 28 single-stranded DNA (ssDNA-comp) overhangs (each was 20 nucleotides (nt’s) in length), which is complementary to the cell-surface-immobilized ssDNA initiators for subsequent nanotile targeting to cells. The bottom surface was decorated with 35 biotin tags for nanotile visualization using fluorescence-conjugated streptavidin. Atomic force microscopy (AFM) imaging of both undecorated and fully decorated DNA nanotiles demonstrated effective origami assembly irrespective of the decoration (Figure 1b, Supplementary Figure 2). This was confirmed by gel electrophoresis (Figure 1c). Our gel electrophoresis studies further confirmed that DNA nanotiles remained stable for 24 h when incubated in PBS (with calcium and magnesium) or Endothelial Cell Growth Media (EGM2) (Supplementary Figure 3).
Figure 1.
Targeting DNA nanotiles to glycocalyx-anchored ssDNA initiators. (a) Diagram showing the design and decoration of the DNA nanotile with ssDNA-comp overhangs and biotin tags. (b) AFM images of assembled DNA nanotiles without decoration and with full decorations (28 ssDNA-comp overhangs and 35 biotin tags). (c) DNA gel electrophoresis analysis of 2-Log DNA ladder, M13 scaffold, undecorated nanotiles, and fully decorated nanotiles. (d) Azide ligands were metabolically incorporated into glycans within the glycocalyx by administering Ac4ManNAz. These cell-surface azide ligands were conjugated with azide-reactive 5′DBCO-ssDNA initiators, leading to covalent immobilization of ssDNA initiators onto the glycocalyx, which can then recruit DNA nanotiles via hybridization with the complementary ssDNA-comp overhangs on nanotiles. (e–g) ssDNA initiators, immobilized on the glycocalyx, were detected through its hybridization with the fluorescent, complementary 5′FAM-ssDNA-comp oligos (e). FAM fluorescence intensity was visualized using microscopic imaging (f) and quantified using a spectrometer (g). (h, i) DNA nanotiles with 35 biotin tags and 1, 3, 6, 14, or 28 complementary ssDNA-comp overhangs were targeted to glycocalyx-anchored ssDNA initiators. Cell-surface nanotiles were visualized via biotin detection using a fluorophore-conjugated streptavidin (red) (h), and the fluorescence intensity was quantified using a spectrometer (i). Data represent means ± s.d. from three independent replicates. **P ≤ 0.01, ***P ≤ 0.001.
To anchor DNA nanotiles directly onto the cell-surface glycocalyx, using human umbilical vein endothelial cells (HUVECs) as a model, we investigated DNA nanotile targeting via the hybridization of glycocalyx-anchored ssDNA initiators with nanotiles bearing complementary ssDNA overhangs. To install the 20-nt ssDNA initiators onto the cell-surface glycocalyx, we employed bioorthogonal glycocalyx labeling with the copper-free click chemistry.44−46 We first incorporated azide ligands covalently onto the glycocalyx through metabolic glycan labeling using an azido monosaccharide, N-azidoacetylmannosamine-tetraacylated (Ac4ManNAz) (Figure 1d and Supplementary Figure 4).46 In parallel, we labeled the 5′-end of ssDNA initiators with dibenzocyclooctyne (DBCO) to generate 5′DBCO-ssDNA and quantified the labeling efficiency to be over 90% using a click shift assay (Supplementary Figure 5).47 This assay detects 5′DBCO-ssDNA via its conjugation with a PEG-azide (10 kDa) and the corresponding increase in molecular weight. Conjugation between azide ligands on the glycocalyx with 5′DBCO-ssDNA led to covalent anchorage of ssDNA initiators onto the glycocalyx (Figure 1d), which was demonstrated by their hybridization with the complementary ssDNA-comp oligos bearing a 5′FAM fluorescent tag (5′FAM-ssDNA-comp) (Figure 1e–g).
We then targeted DNA nanotiles to ssDNA initiators anchored to the cell-surface glycocalyx (Figure 1d). Each DNA nanotile contains 35 biotin tags on its bottom surface and different numbers (1, 3, 6, 14, and 28) of ssDNA-comp overhangs that are evenly spaced across its top surface (Supplementary Figure 6 and Supplementary Figure 7). Biotin decoration of DNA nanotiles offers excellent solubility and allows specific detection of cell-surface-immobilized nanotiles through the staining of cell-impermeable streptavidin. We investigated the quantitative relationship between the number of overhangs per nanotile and its cell-surface immobilization efficiency reflected by the fluorescence intensity of biotin staining using fluorophore-conjugated streptavidin. Gradual increase in the number of ssDNA-comp overhangs per nanotile led to an initial enhancement in cell-surface recruitment, which plateaued when the number of overhangs increased beyond 6 per nanotile (Figure 1h,i). Therefore, DNA nanotiles decorated with 6 ssDNA-comp overhangs were used throughout the rest of the study.
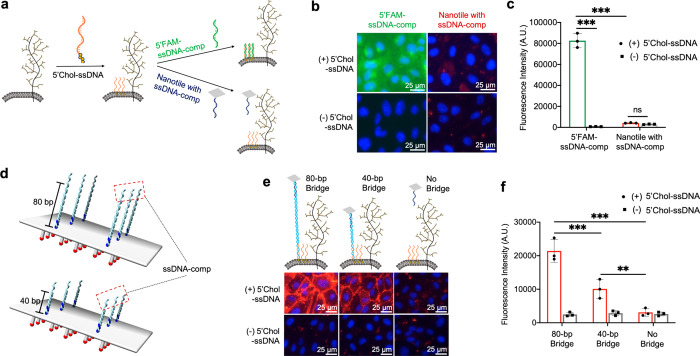
Cholesterol modification, due to its affinity to the PLB, is commonly used for cell-surface targeting of DNA nanostructures.33,48 To investigate how the anchoring mechanisms of ssDNA initiators regulate cell-surface targeting of DNA nanotiles, we compared the performance of ssDNA initiators anchored on the glycocalyx versus those anchored on the PLB. We synthesized a 5′-cholesterol-conjugated ssDNA initiator, referred to as 5′Chol-ssDNA, and verified its PLB targeting using 5′FAM-ssDNA-comp (Figure 2a–c). We then examined if PLB-anchored ssDNA initiators were capable of binding to DNA nanotiles bearing complementary ssDNA-comp overhangs. We found that, although ssDNA initiators on the PLB can effectively recruit free 5′FAM-ssDNA-comp, they failed to recruit DNA nanotiles with ssDNA-comp overhangs (Figure 2a–c). This suggests that ssDNA initiators on the glycocalyx (Figure 1h,i) but not those on the PLB can effectively recruit DNA nanotiles to the cell surface. We speculate that this was because the glycocalyx functioned as a nanoscale barrier that through steric hindrance excluded nanotiles from reaching the underneath PLB. To examine this possibility, we engineered DNA nanotiles with extended nanotile-to-initiator spacing by inserting a DNA duplex bridge between the nanotile and each ssDNA-comp overhang (Figure 2d). We examined DNA duplex bridges with a length of 40 and 80 base pairs (bp’s) and observed length-dependent rescue of nanotile binding to PLB-anchored initiators (Figure 2e,f). This implies that DNA nanotiles but not DNA duplexes can be effectively expelled by the cell-surface glycocalyx. Furthermore, we assessed glycocalyx- and PLB-oriented nanotile targeting to two additional cell types, the Chinese hamster ovary (CHO) cells and adenocarcinomic human alveolar basal epithelial (A549) cells, and observed consistent results compared to HUVECs (Supplementary Figure 8, Supplementary Figure 9), demonstrating the wide applicability of our findings.
Figure 2.
Targeting DNA nanotiles to PLB-anchored ssDNA initiators. (a) 5′Chol-ssDNA initiators were anchored onto the PLB via hydrophobic interaction and used to recruit either 5′FAM-ssDNA-comp oligos or DNA nanotiles bearing ssDNA-comp overhangs. (b, c) Detection of the recruited 5′FAM-ssDNA-comp oligos and DNA nanotiles was performed via fluorescence imaging of FAM and biotin staining (b) and fluorescence quantification using a spectrometer (c). (d) Diagram of inserting DNA duplex bridges of 40 and 80 bp’s between the DNA nanotile and each ssDNA-comp overhang. (e, f) Biotin-based imaging (e) and fluorescence quantification (f) of DNA nanotile recruitment to PLB-anchored ssDNA initiators in the presence or absence of the bridges. Data represent means ± s.d. from three independent replicates. **P ≤ 0.01, ***P ≤ 0.001.
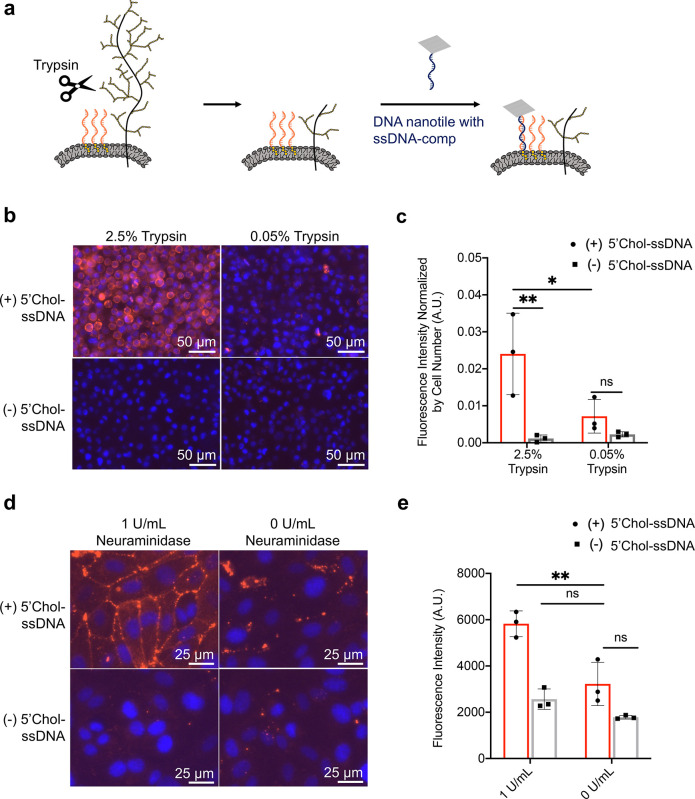
To further verify whether it was the physiologic glycocalyx that expelled DNA nanotiles from binding to PLB-anchored ssDNA initiators, we explored enzymatic degradation of the glycocalyx proteins (Figure 3a). To do this, HUVECs were treated with an augmented regimen of 2.5% trypsin to digest cell-surface proteins, an essential constituent of the glycocalyx, and thereby compromise the glycocalyx barrier integrity. As a control, cells were treated with mild trypsin (0.05%) that is commonly used for cell dissociation. Through fluorescence-based biotin staining, imaging, and quantification, we observed trypsin-dose-dependent enhancement of nanotile recruitment to PLB-anchored ssDNA initiators in the absence of extended nanotile-to-initiator spacing (Figure 3b,c). This confirms that the glycocalyx acted as a nanoscale cell-surface barrier that excluded DNA nanotiles from reaching the underneath PLB.
Figure 3.
Targeting DNA nanotiles to PLB-anchored ssDNA initiators following degradation of glycocalyx. (a) Diagram showing our hypothesis of permitted access of DNA nanotile to the PLB following enzymatic digestion of the glycocalyx via trypsin. HUVECs were pretreated with high (2.5%) or low (0.05%) concentrations of trypsin before the sequential binding of 5′Chol-ssDNA initiators and DNA nanotiles. (b, c) Evaluation of nanotile binding to PLB-anchored initiators by fluorescent streptavidin detection of biotin (red, b) and fluorescence quantification (c). (d, e) HUVECs were pretreated with 1 U/mL neuraminidase for 1 h before the sequential binding of 5′Chol-ssDNA initiators and DNA nanotiles. Fluorescence detection (d) and quantification (e) of DNA nanotile recruitment to PLB-anchored ssDNA initiators via fluorescent streptavidin with and without neuraminidase treatment. Data represent means ± s.d. from three independent replicates. *P ≤ 0.05, **P ≤ 0.01.
Neuraminidase is a glycocalyx-degrading enzyme that cleaves sialic acid residues expressed on cell-surface glycoproteins and glycolipids.42,49 Upregulation of neuraminidase plays a vital role in a variety of pathological conditions, such as atrial stiffening, type 2 diabetes, and viral infections.42,50 Here we examined whether our DNA-nanotile-based PLB accessibility strategy had sufficient sensitivity to detect glycocalyx damage caused by neuraminidase treatment, which is conventionally detected using transmission electron microscopy (Supplementary Figure 10). We targeted the nanotiles to PLB-anchored ssDNA initiators in HUVECs with and without pretreatment of 1 U/mL neuraminidase (1 h).51 Nanotiles without DNA duplex bridges were unable to access PLB-anchored initiators in healthy HUVECs but were able to effectively reach and get immobilized on the surface of HUVECs injured by neuraminidase (Figure 3d,e). Our results confirm that the physiologic glycocalyx acted as a nanoscale cell-surface barrier that excluded DNA nanotiles from reaching the underneath PLB and that the DNA nanotiles can successfully predict the disease-related compromise of glycocalyx barrier integrity.
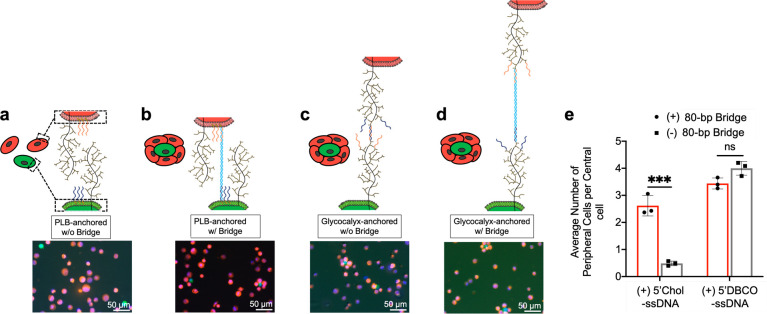
From the cellular engineering perspective, it is of particular interest to understand how cell-surface accessibility regulates the adhesion and the assembly between two groups of cells with surface decoration of mutually complementary ssDNA and ssDNA-comp oligos.44,52,53 As described above, we observed that nanotile recruitment to the cell surface is regulated by ssDNA-initiator-anchoring mechanisms. Here, we investigated its correlation with cell-to-cell accessibility in DNA-guided multicellular assembly.44,52 We mixed at a 1:100 ratio of two groups of color-coded HUVECs, with surface decoration of ssDNA (red cells) and complementary ssDNA-comp (green cells) initiators, respectively, and examined the formation of a red-cell-centered multicellular assembly as a readout of cell-to-cell accessibility (Figure 4). Consistent with the cell-surface accessibility to DNA nanotiles (Figure 1h,i), glycocalyx-anchored ssDNA and ssDNA-comp initiators promoted effective assembly of two-colored cell clusters with desired organization (Figure 4c), while the PLB-anchored initiators could not (Figure 4a). We previously demonstrated that the poor PLB-to-nanotile accessibility can be rescued by double-stranded-DNA (dsDNA)-mediated spacing (Figure 2d–f). To assess whether such spacing modulates the assembly process, the green cells were decorated with a new ssDNA(bridge) initiator, followed by hybridization with the 80-bp dsDNA bridge bearing ssDNA-comp and ssDNA(bridge)-comp (complementary to the ssDNA(bridge) initiator) on both ends, which effectively extended the spacing between the PLB and the ssDNA-comp initiator by 80 bp’s. In parallel, the ssDNA initiator of the red cells remained directly anchored on the PLB without any further spacing. Indeed, the dsDNA-bridge-mediated spacing in the green cells enabled effective assembly of cells bearing PLB-anchored initiators (Figure 4b,e). This is again consistent with DNA-duplex-bridge-mediated control of nanotile-to-PLB accessibility (Figure 2d–f). In parallel, the bridge-mediated spacing did not obviously alter the assembly process mediated by glycocalyx-anchored initiators (Figure 4d,e). These results imply that the outcome of particular cell-surface initiator configurations (anchoring mechanism and spacer length) for cell–cell accessibility in multicellular assembly mirrors its outcome for nanotile-to-cell accessibility. Thus, we envision that DNA nanotiles can be used to assess and optimize the overall cell-surface accessibility prior to performing multicellular assembly in complex tissue and cell engineering applications.
Figure 4.
Cell-to-cell accessibility assessed by multicellular assembly driven by cell-surface ssDNA and ssDNA-comp. Two groups of color-coded HUVECs with surface decoration of mutually complementary ssDNA and ssDNA-comp were mixed at a ratio of 1:100 (green:red) for DNA-guided multicellular assembly. (a, b) Assembly between cell groups with PLB-anchored ssDNA and ssDNA-comp initiators in the absence (a) and presence (b) of the DNA duplex bridge on the green cells. (c, d) Assembly between cell groups with glycocalyx-anchored ssDNA and ssDNA-comp initiators in the absence (c) and presence (d) of the DNA duplex bridge on the green cells. (e) Quantification of the number of peripheral cells per central cell in the resulting assemblies. Data represent means ± s.d. from three independent replicates. ***P ≤ 0.001.
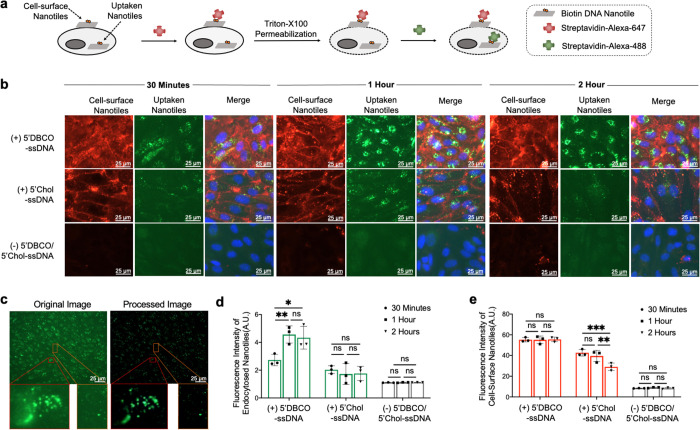
The desirable features of DNA origami, such as biocompatibility and 3D programmability, make DNA origami an emerging platform for intracellular drug delivery.54−56 Following demonstrating glycocalyx- and PLB-oriented mechanisms for targeting DNA origami nanotiles to the cell surface, next we investigated and compared the stability of the resulting cell-surface-immobilized nanotiles. Cells bearing ssDNA initiators on their surface were incubated with DNA nanotiles for 30 min, 1 h, or 2 h at 37 °C, which generated more robust labeling compared to incubation at 4 °C (Supplementary Figures 11 and 12). Following each incubation period, cells were fixed, and the nanotiles remaining on the cell surface and those that had been uptaken by cells were visualized sequentially using a two-step, dual-color staining assay using fluorescence-labeled, cell-impermeable streptavidin. In the first step, the far-red-colored streptavidin (Alexa 647) was introduced to label the nanotiles bound to the external cell surface. In the second step, cells were permeabilized with Triton-X100, and the green-colored streptavidin (Alexa 488) was administered to label intracellular nanotiles that have been uptaken and therefore escaped with the first round of streptavidin (Alexa 647) binding (Figure 5a,b).
Figure 5.
Cellular uptake and stability of cell-surface-anchored DNA nanotiles. (a) Strategy for the two-step, dual-color streptavidin staining of cell-surface and uptaken nanotiles. (b) Dual-color detection of cell-surface (red) and uptaken (green) nanotiles following 30 min, 1 h, or 2 h of incubation. Nanotiles were targeted to the cell surface via glycocalyx- or PLB-anchored initiators. (c) Original and postprocessed fluorescence images of the uptaken nanotiles. (d, e) Fluorescence quantification of the uptaken nanotiles (d) and those remaining on the cell surface (e) over time. Cells incubated with nanotiles in the absence of cell-surface initiators served as the control. Data represent means ± s.d. from three independent replicates. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Next, the uptaken DNA nanotiles were quantified by measuring the signal intensity of the intracellular labeling by streptavidin (Alexa 488). An image processing pipeline was developed to facilitate background subtraction and signal identification (Figure 5c). Comparing the two different nanotile-anchoring mechanisms, glycocalyx anchoring led to enhanced cellular uptake at 30 min, which further increased at 1 h and reached a plateau afterward (Figure 5b,d). In contrast, PLB anchoring resulted in less cellular uptake at 30 min, which did not further increase over time (Figure 5b,d). To investigate the potential mechanism underlying this difference, we quantified the stability of nanotiles located at the external cell surface and observed superior stability of those anchored on the glycocalyx over the 2 h period of investigation (Figure 5b,e). In contrast, the abundance of nanotiles anchored directly on the PLB via cholesterol decreased over time (Figure 5b,e). This is likely because the hydrophobic interaction between the PLB and 5′Chol-ssDNA initiator is non-covalent and reversible, while the glycocalyx-targeted initiator anchoring is covalent in nature.
DNA origami has emerged as a powerful nanotechnology platform for sensing and modulating cellular activities.33,37,38,53,57−60 It is therefore of particular interest to target DNA origami nanostructures directly to the cell surface. Cholesterol labeling has been commonly used for achieving such a purpose,33,61 which directs origami nanostructures to the phospholipid bilayer (PLB). Although cell-surface glycoconjugates have been widely reported and used for oligonucleotide attachment,44,45 it has not been explored for DNA origami targeting. Here we explored this possibility and compared it with the commonly utilized cholesterol-mediated cell targeting, finding that DNA nanostructures anchored directly on the glycocalyx (via the glycoconjugates) exhibited several unique features in terms of accessibility to the cell surface, sensitivity to steric hindrance from the glycocalyx itself, cell-surface stability, and cellular uptake activity. We expect these findings to expand the toolbox for cellular targeting of DNA nanostructures, in particular regarding improving cell-surface and intracellular delivery.
Barrier formation at the tissue level is an essential mechanism that prevents uncontrolled passage of molecules, particles, cells, and microbes across tissue boundaries.1,3,4 This is observed in the endothelium lining the vasculature and epithelium lining the lung and intestinal lumen.5−12 Barrier-forming cells not only establish paracellular junctions, such as tight junctions and adherent junctions, but also use selective deposition of glycocalyx at their luminal surface as a critical apparatus controlling barrier permeability.1,3,4 Aberrant shedding of the luminal glycocalyx is involved in a broad range of pathological conditions, such as atherosclerosis, stroke, hypertension, infection, and inflammation.10,11,15−20,24,25 Despite the critical importance of the glycocalyx in maintaining barrier homeostasis, most current approaches for glycocalyx analysis characterize its morphology rather than its barrier function.26−31 Here we established DNA origami nanotiles as an effective and sensitive measure of the minimal thickness of the glycocalyx barrier. The PLB accessibility of nanotiles is quantitatively regulated by the glycocalyx integrity and by the length of nanotile-to-PLB spacing. The compromised glycocalyx barrier integrity linked to neuraminidase-related diseases was well captured by the PLB accessibility assay of nanotiles in our work. Given the desirable features, such as nanoscale resolution, manufacturing reproducibility, and solubility, the DNA origami is an ideal candidate for future development of probes to monitor the glycocalyx barrier integrity both in vitro and in vivo.
Besides its contribution to tissue boundary formation, the glycocalyx coating also regulates cell-to-cell adhesion. Cell-surface decoration of mutually complementary ssDNA oligos has been widely used to guide programmed assembly of dissociated cells via DNA hybridization. Both glycocalyx- and PLB-anchored ssDNA oligos have been used to guide multicellular assembly with desired cellular composition and spatial arrangement.44,52 However, there has been a lack of experimentally tractable systems to allow mechanistic interpretation of the varied assembly efficiency. Here we showed that the binding of DNA nanotiles to cell-surface ssDNA initiators faithfully predicted the ability of these ssDNA initiators to mediate cell-to-cell assembly, and therefore offered a quantitative means to optimize ssDNA-anchoring mechanisms and spacing to fine-tune multicellular assembly. We also expect our finding to be useful for developing cell therapy applications where cell-to-tissue accessibility is of critical importance.
With the wide range of cargo versatility and programmability, DNA origami is being actively pursued for intracellular drug delivery.62−64 Cholesterol-based cell-surface targeting has recently been used for promoting cellular uptake of DNA nanostructures.48,65 Here we showed that, compared to the cholesterol-directed mechanism, glycocalyx-based DNA nanotile targeting exhibited not only enhanced cell-surface stability but also augmented cellular uptake efficiency. Our glycocalyx-based DNA origami targeting utilizes metabolic azide labeling of glycoproteins within the glycocalyx followed by installation of ssDNA initiators via the click chemistry conjugation. The feasibility of such metabolic azide labeling and bioorthogonal conjugation has been well documented in cell, tissue, and live organism applications.46,47,66 Therefore, our finding offers an alternative option for cellular targeting of the DNA origami with potential applicability both in vitro and in vivo.
In conclusion, our results present compelling evidence that establishes DNA origami nanotiles as a nanoscale functional measure of the glycocalyx barrier integrity. Our study enables future development of DNA-origami-based nanosensors to monitor glycocalyx integrity during dynamic pathophysiological processes. We also offer an expanded toolbox for cell-surface targeting using DNA origami to modulate intercellular and intracellular activities.
Acknowledgments
We are grateful to Drs. Adam W. Feinberg, Philip R. LeDuc, and Rachelle N. Palchesko for sharing the AFM, to Dr. Donna B. Stolz for assistance on transmission electron microscopy, and to Misti West for laboratory management.
Glossary
Abbreviations
- PLB
phospholipid bilayer
- TEM
transmission electron microscopy
- ssDNA
single-stranded DNA (ssDNA)
- nt
nucleotide
- AFM
atomic force microscopy
- HUVECs
human umbilical vein endothelial cells
- Ac4ManNAz
N-azidoacetylmannosamine-tetraacylated
- DBCO
dibenzocyclooctyne
- bp’s
base pairs
- dsDNA
double-stranded DNA
- RT
room temperature
- DBCO-sulfo-NHS-ester
DBCO-sulfo-N-hydroxysuccinimidyl ester
- EGM-2
Endothelial Cell Growth Medium-2
- PFA
paraformaldehyde
- ANOVA
one-way analysis of variance
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c01236.
Supplementary Figures 1–12, Supplementary Tables 1–4, and the Materials and Methods (PDF)
Author Contributions
# P.W. and Y.L. contributed equally. All authors contributed to the design of the project, data interpretation, and preparation of the manuscript. P.W. and Y.L. performed the experiments and analyzed the data. W.W. performed image analysis of the cellular uptake of DNA nanotiles. E.K.J. assisted with the multicellular assembly study. M.L.G.S. performed the transmission electron microscopy study.
This work was supported by the Department of Biomedical Engineering (X.R.) and the Department of Mechanical Engineering (R.E.T.) at Carnegie Mellon University, a T32 predoctoral training grant (Biomechanics in Regenerative Medicine, BiRM) from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (NIH) (P.W.), a 2018 Dowd Fellowship (Y.L.), the Samuel and Emma Winters Foundation (R.E.T.), the National Heart Lung and Blood Institute of the NIH 1R21HL152147 (R.E.T.), and the Berkman Faculty Development Fund (R.E.T.).
The authors declare no competing financial interest.
Supplementary Material
References
- Reitsma S.; Slaaf D. W.; Vink H.; van Zandvoort M. A. M. J.; oude Egbrink M. G. A. The endothelial glycocalyx: composition, functions, and visualization. Pfluegers Arch. 2007, 454, 345–359. 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Xie Y.; Wong M.; Lebrilla C. B. Characterization of Cell Glycocalyx with Mass Spectrometry Methods. Cells 2019, 8, 882. 10.3390/cells8080882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Chu E. R.; Malafeev A.; Pajarskas T.; Pivkin I. V.; Koumoutsakos P. Structure and response to flow of the glycocalyx layer. Biophys. J. 2014, 106, 232–243. 10.1016/j.bpj.2013.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbaum S.; Tarbell J. M.; Damiano E. R. The Structure and Function of the Endothelial Glycocalyx Layer. Annu. Rev. Biomed. Eng. 2007, 9, 121–167. 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- Henry C. B.; Duling B. R. Permeation of the luminal capillary glycocalyx is determined by hyaluronan. Am. J. Physiol. 1999, 277, H508–514. 10.1152/ajpheart.1999.277.2.H508. [DOI] [PubMed] [Google Scholar]
- Lipowsky H. H. The endothelial glycocalyx as a barrier to leukocyte adhesion and its mediation by extracellular proteases. Ann. Biomed. Eng. 2012, 40, 840–848. 10.1007/s10439-011-0427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. P.; et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat. Med. 2012, 18, 1217–1223. 10.1038/nm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. B.; Duran W. N.; DeFouw D. O. Permselectivity of angiogenic microvessels following alteration of the endothelial fiber matrix by oligosaccharides. Microvasc. Res. 1997, 53, 150–155. 10.1006/mvre.1996.1996. [DOI] [PubMed] [Google Scholar]
- Dogné S.; Flamion B.; Caron N. Endothelial Glycocalyx as a Shield Against Diabetic Vascular Complications. Arterioscler., Thromb., Vasc. Biol. 2018, 38, 1427–1439. 10.1161/ATVBAHA.118.310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeger S. M.; et al. Epithelial Heparan Sulfate Contributes to Alveolar Barrier Function and Is Shed during Lung Injury. Am. J. Respir. Cell Mol. Biol. 2018, 59, 363–374. 10.1165/rcmb.2017-0428OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venge P.; et al. Subcutaneous administration of hyaluronan reduces the number of infectious exacerbations in patients with chronic bronchitis. Am. J. Respir. Crit. Care Med. 1996, 153, 312–316. 10.1164/ajrccm.153.1.8542136. [DOI] [PubMed] [Google Scholar]
- Frey A.; et al. Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: implications for microbial attachment and oral vaccine targeting. J. Exp. Med. 1996, 184, 1045–1059. 10.1084/jem.184.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arokiasamy S. Heparanase-Dependent Remodeling of Initial Lymphatic Glycocalyx Regulates Tissue-Fluid Drainage During Acute Inflammation in vivo. Front. Immunol. 2019, 10, 2316. 10.3389/fimmu.2019.02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y.; Tarbell J. M. The Adaptive Remodeling of Endothelial Glycocalyx in Response to Fluid Shear Stress. PLoS One 2014, 9, e86249 10.1371/journal.pone.0086249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar A.; et al. Degradation of Glycocalyx and Multiple Manifestations of Endothelial Dysfunction Coincide in the Early Phase of Endothelial Dysfunction Before Atherosclerotic Plaque Development in Apolipoprotein E/Low-Density Lipoprotein Receptor-Deficient Mice. J. Am. Heart Assoc. 2019, 8, e011171 10.1161/JAHA.118.011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.; et al. Glycocalyx in Atherosclerosis-Relevant Endothelium Function and as a Therapeutic Target. Curr. Atheroscler. Rep. 2017, 19, 63–63. 10.1007/s11883-017-0691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaValle B.; et al. Multiple Soluble Components of the Glycocalyx Are Increased in Patient Plasma After Ischemic Stroke. Stroke 2019, 50, 2948–2951. 10.1161/STROKEAHA.119.025953. [DOI] [PubMed] [Google Scholar]
- Ikonomidis I.; et al. Impaired Arterial Elastic Properties and Endothelial Glycocalyx in Patients with Embolic Stroke of Undetermined Source. Thromb. Haemostasis 2019, 119, 1860–1868. 10.1055/s-0039-1694752. [DOI] [PubMed] [Google Scholar]
- Ikonomidis I.; et al. Association of impaired endothelial glycocalyx with arterial stiffness, coronary microcirculatory dysfunction, and abnormal myocardial deformation in untreated hypertensives. J. Clin. Hypertens. 2018, 20, 672–679. 10.1111/jch.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafyllidi H.; et al. HDL cholesterol levels and endothelial glycocalyx integrity in treated hypertensive patients. J. Clin. Hypertens. (Hoboken, NJ, U. S.) 2018, 20, 1615–1623. 10.1111/jch.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba T.; Levy J. H. Derangement of the endothelial glycocalyx in sepsis. J. Thromb. Haemostasis 2019, 17, 283–294. 10.1111/jth.14371. [DOI] [PubMed] [Google Scholar]
- Abassi Z.; Armaly Z.; Heyman S. N. Glycocalyx Degradation in Ischemia-Reperfusion Injury. Am. J. Pathol. 2020, 190, 752–767. 10.1016/j.ajpath.2019.08.019. [DOI] [PubMed] [Google Scholar]
- Rehm M.; et al. Shedding of the Endothelial Glycocalyx in Patients Undergoing Major Vascular Surgery With Global and Regional Ischemia. Circulation 2007, 116, 1896–1906. 10.1161/CIRCULATIONAHA.106.684852. [DOI] [PubMed] [Google Scholar]
- Pruessmeyer J.; et al. A disintegrin and metalloproteinase 17 (ADAM17) mediates inflammation-induced shedding of syndecan-1 and −4 by lung epithelial cells. J. Biol. Chem. 2010, 285, 555–564. 10.1074/jbc.M109.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chignalia A. Z.; et al. The glycocalyx and Trauma: A Review. Shock 2016, 45, 338–348. 10.1097/SHK.0000000000000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell D.; et al. The Glycocalyx of the Human Umbilical Vein Endothelial Cell: An Impressive Structure Ex Vivo but Not in Culture. Circ. Res. 2009, 104, 1313–1317. 10.1161/CIRCRESAHA.108.187831. [DOI] [PubMed] [Google Scholar]
- Ebong E. E.; Macaluso F. P.; Spray D. C.; Tarbell J. M. Imaging the Endothelial Glycocalyx In Vitro by Rapid Freezing/Freeze Substitution Transmission Electron Microscopy. Arterioscler., Thromb., Vasc. Biol. 2011, 31, 1908–1915. 10.1161/ATVBAHA.111.225268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B. M.; Vink H.; Spaan J. A. E. The Endothelial Glycocalyx Protects Against Myocardial Edema. Circ. Res. 2003, 92, 592–594. 10.1161/01.RES.0000065917.53950.75. [DOI] [PubMed] [Google Scholar]
- Luft J. H. Fine structures of capillary and endocapillary layer as revealed by ruthenium red. Fed. Proc. 1966, 25, 1773–1783. [PubMed] [Google Scholar]
- Florian J. A.; et al. Heparan sulfate proteoglycan is a mechanosensor on endothelial cells. Circ. Res. 2003, 93, e136–e142. 10.1161/01.RES.0000101744.47866.D5. [DOI] [PubMed] [Google Scholar]
- Mulivor A. W.; Lipowsky H. H. Inflammation- and ischemia-induced shedding of venular glycocalyx. American Journal of Physiology-Heart and Circulatory Physiology 2004, 286, H1672–H1680. 10.1152/ajpheart.00832.2003. [DOI] [PubMed] [Google Scholar]
- Vink H.; Duling B. R. Identification of distinct luminal domains for macromolecules, erythrocytes, and leukocytes within mammalian capillaries. Circ. Res. 1996, 79, 581–589. 10.1161/01.RES.79.3.581. [DOI] [PubMed] [Google Scholar]
- Akbari E.; et al. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29, 1703632. 10.1002/adma.201703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Andersen E. S.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney C. J.; Lucas C. R.; O’Brien F. J.; Castro C. E. DNA Origami: Folded DNA-Nanodevices That Can Direct and Interpret Cell Behavior. Adv. Mater. 2016, 28, 5509–5524. 10.1002/adma.201504733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Arias D. S.; Deserno M.; Ren X.; Taylor R. E. Emerging Applications at the Interface of DNA Nanotechnology and Cellular Membranes: Perspectives from Biology, Engineering and Physics. APL Bioengineering 2020, 4, 041507. 10.1063/5.0027022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote C. A. The targeted inhibition of neuraminidase reverses endothelial glycocalyx degradation and improves endothelial function in type 2 diabetes. FASEB J. 2019, 33, 527.16. 10.1096/fasebj.2019.33.1_supplement.527.16. [DOI] [Google Scholar]
- Foote C. A.; et al. Arterial Stiffening in Western Diet-Fed Mice Is Associated with Increased Vascular Elastin, Transforming Growth Factor-beta, and Plasma Neuraminidase. Front. Physiol. 2016, 7, 285. 10.3389/fphys.2016.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieve I.; et al. A positive feedback loop between IL-1beta, LPS and NEU1 may promote atherosclerosis by enhancing a pro-inflammatory state in monocytes and macrophages. Vasc. Pharmacol. 2018, 103–105, 16–28. 10.1016/j.vph.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Palese P.; Tobita K.; Ueda M.; Compans R. W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 1974, 61, 397–410. 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- Basak S.; Tomana M.; Compans R. W. Sialic-Acid Is Incorporated into Influenza Hemagglutinin Glycoproteins in the Absence of Viral Neuraminidase. Virus Res. 1985, 2, 61–68. 10.1016/0168-1702(85)90060-7. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Bertozzi C. R. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 4606–4610. 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R. A.; Douglas E. S.; Mathies R. A.; Bertozzi C. R.; Francis M. B. Programmable Cell Adhesion Encoded by DNA Hybridization. Angew. Chem., Int. Ed. 2006, 45, 896–901. 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- Saxon E.; Bertozzi C. R. Cell Surface Engineering by a Modified Staudinger Reaction. Science 2000, 287, 2007–2010. 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- Ren X.; et al. Metabolic glycan labeling and chemoselective functionalization of native biomaterials. Biomaterials 2018, 182, 127–134. 10.1016/j.biomaterials.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Whitehouse W. L.; Noble J. E.; Ryadnov M. G.; Howorka S. Cholesterol Anchors Enable Efficient Binding and Intracellular Uptake of DNA Nanostructures. Bioconjugate Chem. 2019, 30, 1836–1844. 10.1021/acs.bioconjchem.9b00036. [DOI] [PubMed] [Google Scholar]
- Gottschalk A. The influenza virus neuraminidase. Nature 1958, 181, 377–378. 10.1038/181377a0. [DOI] [PubMed] [Google Scholar]
- Air G. M.; Laver W. G. The neuraminidase of influenza virus. Proteins: Struct., Funct., Genet. 1989, 6, 341–356. 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- Barker A. L.; et al. Observation and characterisation of the glycocalyx of viable human endothelial cells using confocal laser scanning microscopy. Phys. Chem. Chem. Phys. 2004, 6, 1006–1011. 10.1039/B312189E. [DOI] [Google Scholar]
- Teramura Y.; Chen H.; Kawamoto T.; Iwata H. Control of cell attachment through polyDNA hybridization. Biomaterials 2010, 31, 2229–2235. 10.1016/j.biomaterials.2009.11.098. [DOI] [PubMed] [Google Scholar]
- Ge Z.; et al. Programming Cell-Cell Communications with Engineered Cell Origami Clusters. J. Am. Chem. Soc. 2020, 142, 8800–8808. 10.1021/jacs.0c01580. [DOI] [PubMed] [Google Scholar]
- Jiang D.; et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nature Biomedical Engineering 2018, 2, 865–877. 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q.; Liu S.; Liu J.; Wang Z.-G.; Ding B. Rationally Designed DNA-Origami Nanomaterials for Drug Delivery In Vivo. Adv. Mater. 2019, 31, 1804785. 10.1002/adma.201804785. [DOI] [PubMed] [Google Scholar]
- Tasciotti E. Smart cancer therapy with DNA origami. Nat. Biotechnol. 2018, 36, 234–235. 10.1038/nbt.4095. [DOI] [PubMed] [Google Scholar]
- Bastings M. M. C.; et al. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18, 3557–3564. 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- Liu S.; et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 2021, 20, 421–430. 10.1038/s41563-020-0793-6. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy N.; et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa T.; et al. DNA Origami Scaffolds as Templates for Functional Tetrameric Kir3 K(+) Channels. Angew. Chem., Int. Ed. 2018, 57, 2586–2591. 10.1002/anie.201709982. [DOI] [PubMed] [Google Scholar]
- Johnson-Buck A.; Jiang S.; Yan H.; Walter N. G. DNA-Cholesterol Barges as Programmable Membrane-Exploring Agents. ACS Nano 2014, 8, 5641–5649. 10.1021/nn500108k. [DOI] [PubMed] [Google Scholar]
- Lee H.; et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012, 7, 389–393. 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh A. S.; Yin H.; Erben C. M.; Wood M. J.; Turberfield A. J. DNA cage delivery to mammalian cells. ACS Nano 2011, 5, 5427–5432. 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643. 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- Akbari E. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29, 1703632. 10.1002/adma.201703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. V.; et al. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1821–1826. 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.