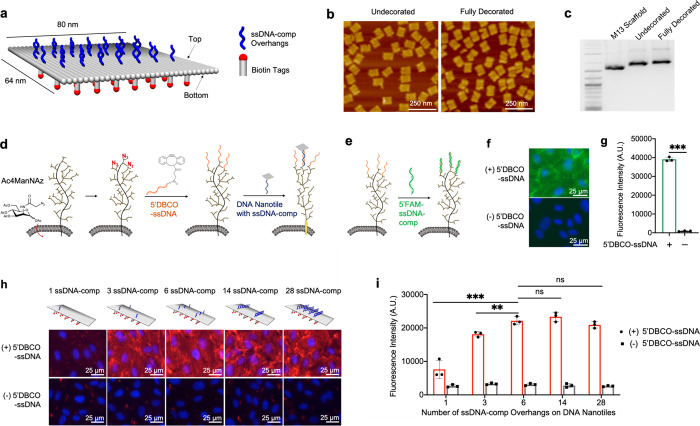
Figure 1.
Targeting DNA nanotiles to glycocalyx-anchored ssDNA initiators. (a) Diagram showing the design and decoration of the DNA nanotile with ssDNA-comp overhangs and biotin tags. (b) AFM images of assembled DNA nanotiles without decoration and with full decorations (28 ssDNA-comp overhangs and 35 biotin tags). (c) DNA gel electrophoresis analysis of 2-Log DNA ladder, M13 scaffold, undecorated nanotiles, and fully decorated nanotiles. (d) Azide ligands were metabolically incorporated into glycans within the glycocalyx by administering Ac4ManNAz. These cell-surface azide ligands were conjugated with azide-reactive 5′DBCO-ssDNA initiators, leading to covalent immobilization of ssDNA initiators onto the glycocalyx, which can then recruit DNA nanotiles via hybridization with the complementary ssDNA-comp overhangs on nanotiles. (e–g) ssDNA initiators, immobilized on the glycocalyx, were detected through its hybridization with the fluorescent, complementary 5′FAM-ssDNA-comp oligos (e). FAM fluorescence intensity was visualized using microscopic imaging (f) and quantified using a spectrometer (g). (h, i) DNA nanotiles with 35 biotin tags and 1, 3, 6, 14, or 28 complementary ssDNA-comp overhangs were targeted to glycocalyx-anchored ssDNA initiators. Cell-surface nanotiles were visualized via biotin detection using a fluorophore-conjugated streptavidin (red) (h), and the fluorescence intensity was quantified using a spectrometer (i). Data represent means ± s.d. from three independent replicates. **P ≤ 0.01, ***P ≤ 0.001.