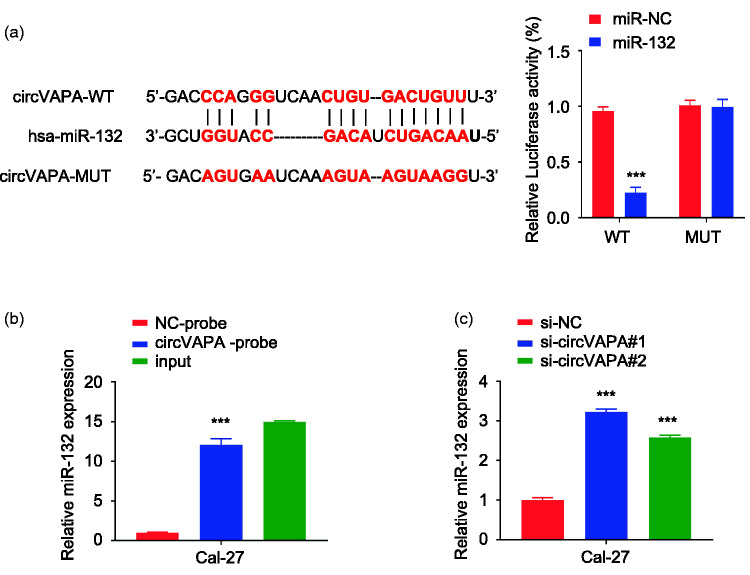
Figure 3.
CircVAPA targets miR-132. a: The online StarBase database predicted a miR-132 binding site in the 3ʹ-untranslated region (UTR) of wildtype (WT) circVAPA. This site was then mutated (MUT). Luciferase reporter experiments were carried out in Cal-27 cells transfected with a negative control (NC) or miR-132. ***P < 0.001 compared with miR-NC. b: The RNA pull-down assay with a biotin-labeled circVAPA probe confirmed that circVAPA directly interacted with miR-132. Input=10% of total lysate. ***P < 0.001 compared with NC-probe. c: RT-qPCR was used to detect the expression of miR-132 in Cal-27 cells. ***P < 0.001 compared with si-NC