Abstract
Background
Dietary content and environmental factors can shape the gut microbiota, and consequently, the way the gut microbiota metabolizes fats, carbohydrates, and proteins, affecting overall health of the host. We evaluated the impact of 3 diets (all meat [raw], high-insoluble fiber dry extruded diet and hydrolyzed protein dry extruded diet) on the gut microbiota of healthy dogs in a cross-over sequential study.
Results
We showed that diet can have an effect on the gut microbiome in dogs, which was influenced by the order of feeding. High-protein (all meat) diets were characterized by an increase in bacteria belonging to the Fusobacteria and Bacteroidetes phyla, whereas a high-insoluble fiber commercial diet correlated with increases in Firmicutes and Actinobacteria phyla. However, the individual dog’s baseline microbiota had the most impact on the magnitude and nature of the changes in response to dietary intervention.
Conclusion
Our results suggest that the dog fecal microbiota is driven by protein and fiber composition to different degrees in individual animals, and targeted modification of these patterns could be useful in the modulation of the gut microbiota in different diseases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-021-00101-8.
Keywords: Dog microbiota, Diet, High-insoluble fiber, High-protein, Hydrolyzed, Raw meat diets
Background
The gut microbiota is essential for maintaining health, as it exerts several beneficial effects on the host, regulates numerous biological pathways; and interacts directly and indirectly with various organs and systems in the body, including the brain, liver, bone, and cardiovascular system [1]. The gut microbiota is a highly complex community that evolves rapidly and adapts to its host over a lifetime and exhibits a remarkable plasticity to environmental changes, particularly diet [2, 3].
Even short-term dietary changes have been shown to alter human gut microbiota composition and changes can be observed within 1–3 days when there are extreme dietary modifications such as switching between an all-meat to an all-plant diet [4]. Similar studies have been performed regarding the effect of fiber on the gut microbiota of dogs [5–15]. Some studies have shown beneficial effects of fiber and changes in the gut microbiota [5, 15], whereas others have not shown any significant change [6, 12, 14]. Results have been dependent on the type of fiber, percentage of fiber, previous diet fed, duration of treatment, health status and methodology used during the analysis. The modern pet food industry uses several fiber sources (mainly by-products derived from the processing of grains, fruits, and vegetables) in the formulation of diets for dogs [15, 16]. There is still a paucity of information regarding the effect of many fiber sources on the composition and activity of the intestinal microbiota of dogs and cats. Similarly, studies have also been published assessing the effect of protein [17–19], and recently with emphasis in obesity [20, 21], and raw meat diets in dogs [8, 22, 23], but more studies are needed to understand this complex interaction under different feeding conditions.
Dysbiosis of the intestinal microbiota has been linked to chronic intestinal inflammation in people, dogs, and cats [24–28]. Chronic intestinal diseases in dogs are often treated by dietary modification, aimed at reducing antigenic stimulation to the intestine [29]. Hydrolyzed diets are composed of low molecular weight (MW) proteins and peptides in order to evade the intestinal immune system [30]. In addition, many commercial veterinary hydrolyzed diets will have other alterations (such as increased polyunsaturated fats) compared to standard veterinary diets [31]. Hydrolyzed diets are associated with beneficial changes in the intestinal microbiota and clinical signs of dogs with chronic enteropathies [32, 33]. However, to our knowledge, there is no published data on how hydrolyzed diets affect the gastrointestinal microbiome in healthy dogs fed different types of diets prior to the change, and in turn, which component of the diet is having the most impact.
The aim of this study was to investigate the effects of dietary modification with a high-protein (all meat) diet, a high-insoluble fiber diet and a hydrolyzed diet on the fecal microbiota of healthy dogs in a cross-over trial.
Results
Effect of diet on the relative abundance of bacterial groups
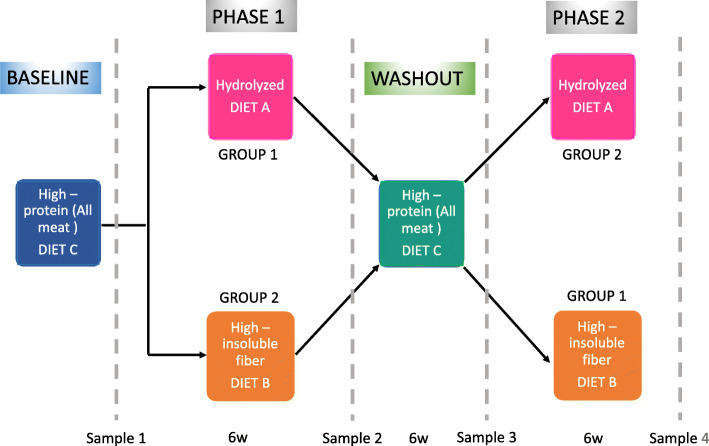
Dogs were classified into two groups. Group 1 dogs were fed diet sequence ACB, and group 2 dogs were fed diet sequence BCA (A = hydrolyzed diet; B = high-insoluble fiber diet; C = high-protein diet [all meat/carcass, raw diet]), each feeding period lasting for 6 weeks. All dogs were fed with a high-protein diet (diet C [all meat/carcass, raw diet]) at baseline (Fig. 1).
Fig. 1.
Schematic design of the cross-over over study. Participant dogs switched diets after a washout period. Sample collection (4 time-points). Each stage of the trial consisted in a 6-week period
The relative abundance of the different bacteria at phylum and family phylogenetic levels were compared among the different categories of diet. At phylum level, Firmicutes, Bacteroidetes and Fusobacteria were the most populous bacterial phyla found, as previously reported [34]. Firmicutes had a median of 44% [range: 18–91%] with the high-protein diet (diet C), a median of 62% [range: 29–93%] with the high-insoluble fiber diet (diet B) and a median of 55% [range: 30–95%] with the hydrolyzed diet (diet A). For Bacteroidetes, the median was 14% [range: 0.22–50%] for the high-protein diet, 16% [range: 0.44–41%] for the high-insoluble fiber diet and 16% [range: 0.34–51%] for the hydrolyzed diet. Meanwhile, for Fusobacteria the median was 24% [range: 4–72%] for the high-protein diet, 8% [range: 1–45%] for the high-insoluble fiber diet and 17% [range: 2–34%] for the hydrolyzed diet (Fig. 2A). The relative abundances at family level are shown in Fig. 2B.
Fig. 2.
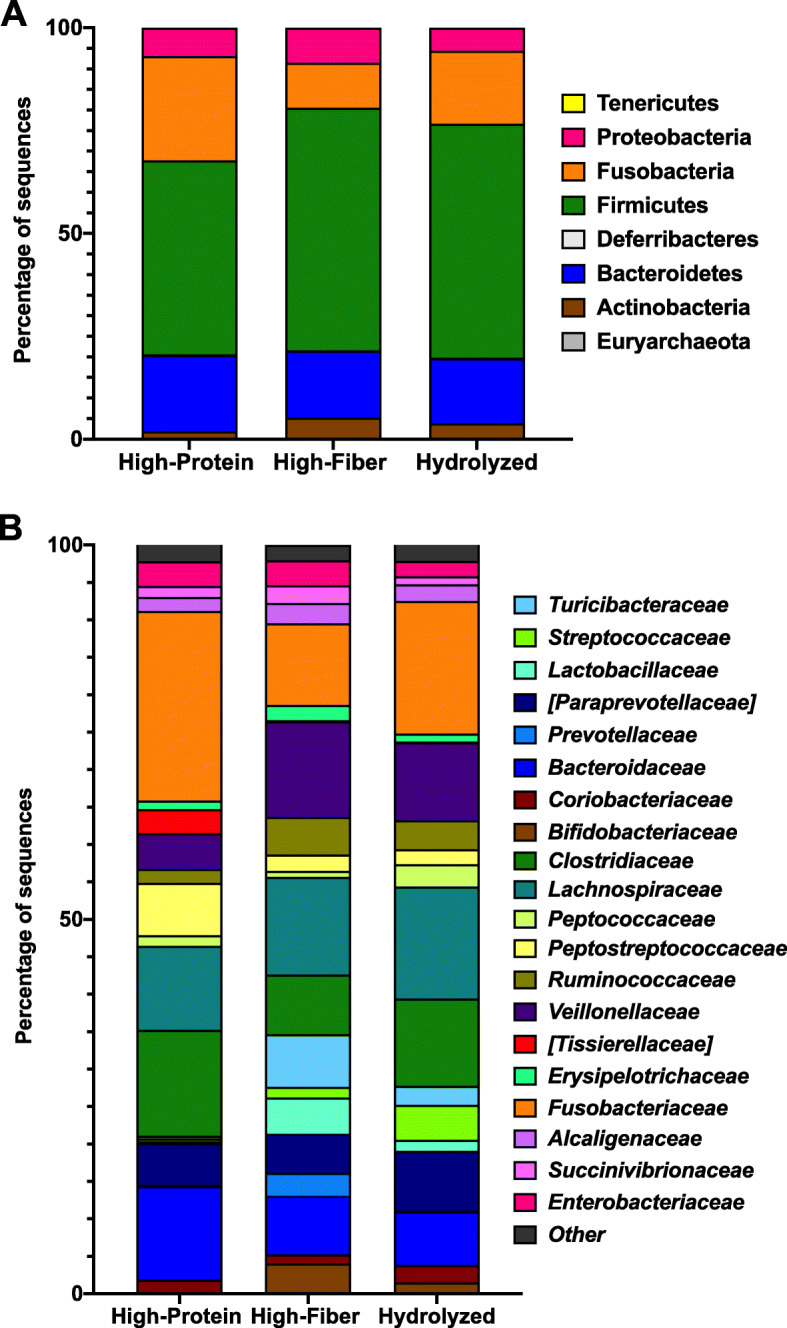
A Relative abundance of bacteria at phylum level in the three diets, irrespective of sequence fed. B Relative abundance of bacteria at family level with the three diets, irrespective of sequence fed. High-Protein (diet C); Hydrolyzed diet (diet A) and High-insoluble fiber (diet B)
Next, we assessed the relative abundance of the different bacterial groups during the baseline and the washout periods (when dogs were being fed diet C- the raw meat/high-protein diet) and found that the relative abundance of some phyla differed between these periods. During baseline, approximately 31% (median) of the sequences corresponded to Bacteroidetes [range: 3–50%], whereas at the end of the washout period, the percentage was 5% [range: 0.22–33%]. For Firmicutes, during baseline the percentage was 37% [range: 18–71%] versus 54% [range: 18–91] during the washout period (Fig. 3). This difference was observed irrespective of the diet sequence for individual dogs.
Fig. 3.
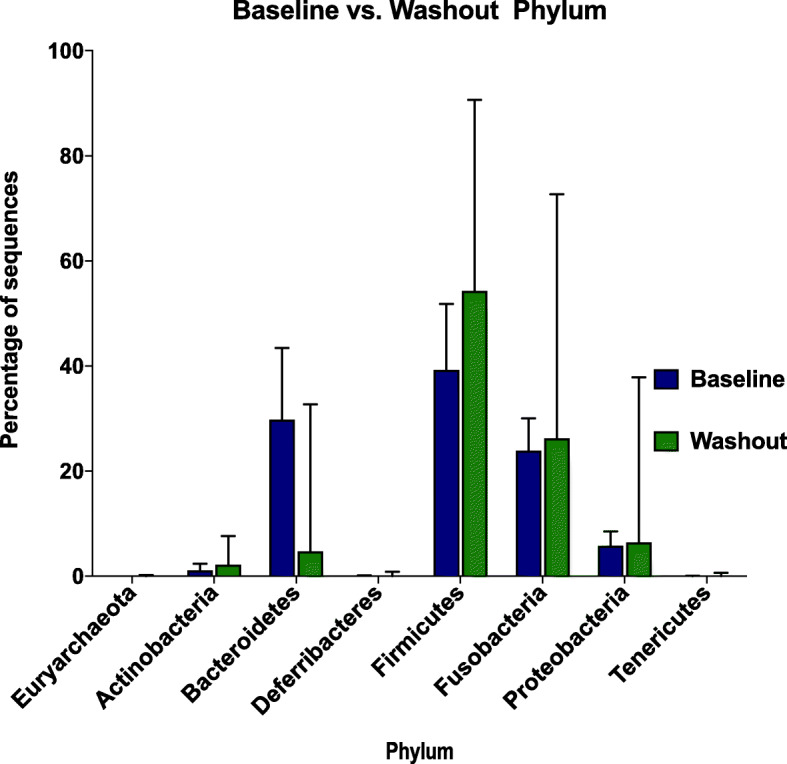
Relative abundance of bacteria at phylum level Baseline versus Washout period. (High-protein diet) (diet C). N = 44, n = 44 each period. Median with range. N: number of dogs. n: number of samples
Analysis of the relative abundance of the different phyla in the hydrolyzed and high-insoluble fiber diets also differed between the ACB and BCA sequences. For example, samples taken from dogs at the end of the 6-week period being fed with the hydrolyzed diet (diet A) showed a relative abundance of Bacteroidetes of 24% (median) [range: 0.71–51] in ACB versus 7% (median) [range: 0.34–29%] in the BCA sequence. However, when the percentages were compared with the preceding diet in each diet sequence; the introduction of the hydrolyzed diet (diet A) did not affect the percentage of Bacteroidetes in any of the diets. In the ACB diet, the percentage of Bacteroidetes ranged between 3 and 50% (median: 23%) at baseline (high-protein, raw meat diet) and for the BCA diet, the percentage ranged between 0.5–33% (median: 8%) at the end of the washout (high-protein, raw diet) period. Thus, changes should be interpreted based on the diet sequence and the preceding microbial profile of each subject, and not independently (See Additional file 1: Fig. S1).
Dietary effects on gut microbial alpha and beta diversity
Alpha diversity was analyzed using the Shannon index considering the subject, as well as the dietary intervention (time point) and diet sequence. In general, Shannon diversity index was not affected by the change of diet when time and subject were considered; although it was lower in the washout period compared to baseline and slightly higher in BCA diet sequence in comparison to ACB diet sequence. The marginal R2 is about 0.3, which suggests that the diet and sequence effects together describe about 30% of the variance in Shannon index [35] (See Additional file 2: Table S1).
In response to the diets, we see a large shift in the overall taxonomic composition of the microbiome. Beta diversity principal-coordinate analysis (PCoA) plots constructed using the Bray-Curtis distance showed a clear separation between high-protein (diet C) with the hydrolyzed diet (diet A) and high-insoluble fiber (diet B) diets (See Additional file 3: Fig. S2A; diet sequence ACB and Additional file 4: Fig. S3A; diet sequence BCA). The hydrolyzed and high-insoluble fiber diets used in our study have some similar nutritional characteristics (e.g., total protein, total carbohydrate) compared to the high-protein diet that could explain, in part, the clustering pattern. Permutational multivariate analysis of variance (PERMANOVA) (Adonis) analysis showed that the type of diet explained ~ 20% of the variability in beta- diversity (R2: 19, p: 0.001), whereas diet sequence only explained 1% of the variability (R2: 1, p: 0.007). When the interaction of these two factors were assessed, diet sequence explained ~ 5% of the variability caused by the type of diet (R2: 6, p: 0.001).
In accordance with the results showed in the relative abundance tables, PERMANOVA (Adonis) analysis using the Bray-Curtis distance identified a significant difference in beta-diversity between the baseline and washout periods (R2: 25, p: 0.001).
Analysis of each group separately, showed that the shifts of the microbiota increased over time when compared to the baseline diet, and was independent of the diet sequence (See Additional file 3: Fig. S2B; diet sequence ACB and Additional file 4: Fig. S3B; diet sequence BCA). In line with the previous clustering pattern, the distance between the hydrolyzed and high-insoluble fiber diet was smaller. The consistency of the community shift argues for a direct effect of the diet as, in the absence of intervention, the dog microbiota has been reported to be stable over time, using 16S rRNA profiling [36].
Differential dietary effects on gut bacterial phyla and families
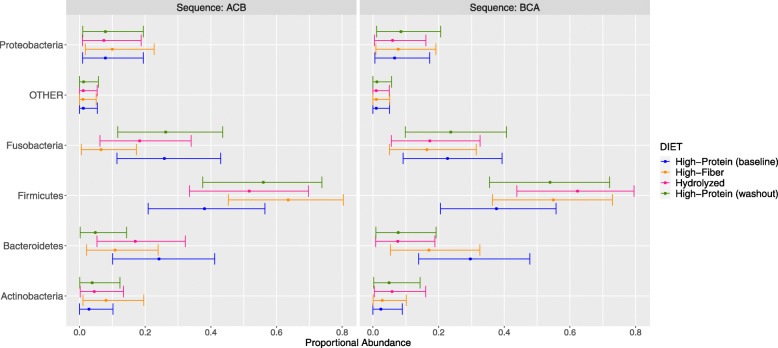
A Dirichlet regression model was performed to compare the microbial differential abundance in each diet sequence considering the variation between dogs and the diets. At phylum level, the high-protein diet (diet C) was enriched with Fusobacteria, for the ACB diet sequence, whereas the high insoluble-fiber (diet B) and hydrolyzed diet (diet A) induced an enrichment in the Firmicutes phylum. Firmicutes was also enriched in the washout period but not during the baseline, whereas Bacteroidetes was enriched only at baseline but not during the washout period. In addition, Actinobacteria was enriched in the high-insoluble fiber diet but only in the ACB sequence (Fig. 4).
Fig. 4.
The posterior estimated predictive distribution of relative abundances at phylum level in ACB and BCA sequence. Points are the posterior population mean. The bars are the 89% prediction intervals from the posterior distribution. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-insoluble Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C)
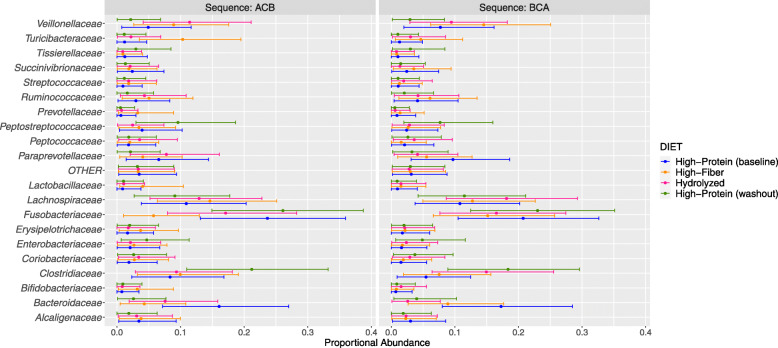
At family level, the 20 most abundant families were assessed. These results were also dependent on the diet sequence, suggesting that the outcome of dietary intervention is influenced by the previous dietary history and the baseline microbiome of the individual. For example, Turicibacteraceae, Lactobacillaceae, Bifidobacteriaceae and Erysipelotrichaceae were more abundant in the high-insoluble fiber samples, but only in the ACB diet sequence. Peptostreptococcaceae and Clostridiaceae were more abundant in the high-protein (diet C) samples only during the washout period, whereas Bacteroidaceae was more abundant only at baseline, and Fusobacteriaceae was more abundant in both periods, regardless of diet sequence. For the hydrolyzed diet, only Veillonellaceae was more abundant in comparison with the other diets, but only during ACB diet sequence (Fig. 5). This family was also more abundant in diet B. Veillonellaceae has been positively correlated with fiber intake [37].
Fig. 5.
The posterior estimated predictive distribution of relative abundances at family level in ACB and BCA sequence. Top of the 20 most abundant families. Points are the posterior population mean. The bars are the 89% prediction intervals from the posterior distribution. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-insoluble Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C)
At the genus level, the ratio of Prevotella to Bacteroides has also been found to be important in the human gut microbiome; it changes in response to diet, with higher Prevotella relative abundance being observed with high carbohydrate diets, while higher relative abundance of Bacteroides has been associated with a high-protein diet [38]. In our study, we observed that the ratio of Prevotella to Bacteroides was higher in the hydrolyzed and high-insoluble fiber diets compared to the high-protein diet (See Additional file 5: Fig. S4).
Functional changes in the gut microbiota
Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) was performed on the 16S rRNA gene gut bacterial composition data to predict Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (KOs) and pathways [39]. All predicted KO pathways at L2 level were subjected to a linear mixed-effects model considering the type of diet and the diet sequence as predictors of the effect of each functional pathway. However, there were no clear effects of diet type or sequence on the predicted metagenome functional content.
Discussion
Several studies have been conducted on the effect of diet on the gut microbiota in humans and rodents, and more studies are emerging in dogs. However, the gut microbiota-diet relationship is complex and challenging to characterize as many factors may influence the results [2]. In our study, all dogs were receiving the same baseline diet, were the same breed, similar age, similar body condition and lived in the same environment, which served to eliminate many confounders that could influence the results. In general, we observed that the selected diets had a substantial residual impact on the fecal communities of all dogs, and results were dependent on the composition of the gut microbiota at the start of the intervention.
Analysis of the alpha diversity between the different categories of diet, showed that overall, the high-insoluble fiber diet and hydrolyzed diet have a higher Shannon diversity in comparison with the raw meat, high-protein diet. However, when the analysis was done per subject, the difference in Shannon diversity was minimal between diets.
Studies investigating the direct impact of protein on gut microbiota composition and functionality have shown that protein quality and source are as important as total amounts in people [2], but less so in dogs [40]. Analysis of the gut microbiota showed that diet C (raw all-meat/high-protein diet) in our study was characterized by an overrepresentation of bacteria belonging to the Fusobacteria phylum. This contrasts with a previous study made in obese and lean dogs with high-protein dietary intervention, where the changes in Fusobacteria were relatively small, although the levels of protein differed between studies (49.38% vs. 19.8% in ours) and was of a shorter duration [20]. Another study where the dry commercial diet was changed to minced raw beef, also showed minimal changes in the Fusobacteria content [17]. High levels of Fusobacterium sp. have been observed in multiple carnivore species [3, 41, 42]. In humans, increased Fusobacterium levels are seen in people consuming a diet high in red and processed meats and are associated with an increased risk of development of colorectal cancer [43, 44]. In one study of dogs fed a red meat diet for 9 weeks, an increased Fusobacterium abundance (~ 15%) was present at 6 weeks, suggesting these changes may take time to develop [19]. Diet C in our study consisted mainly of horse carcass, which is a vastly greater percentage of protein in diet than most commercial diets (prescription or supermarket brand). Digestibility of scrap meat may be lower than for high quality protein due to the high amount of connective tissue, and digestibility of macronutrients may also influence the colonic microbiome [45]. Additionally, most commercial dog food does not contain horse protein, which may be biologically different than other sources of protein [46, 47].
At lower phylogenetic levels, an overrepresentation of members of the families Clostridiaceae and Peptostreptococcaceae were also found in the samples from dogs fed diet C. Clostridium is important for lysine and proline utilization by the host, via fermentation in the colon; while Peptostreptococci drives tryptophan and glutamate catabolism [48]. In people, an exclusively meat-based diet is frequently associated with high levels of bile-tolerant bacterial species like Bacteroides and low levels of Prevotella [38]. Of interest, in our study, Clostridiaceae and Peptostreptococcaceae were only enriched in the washout period, whereas the Bacteroidaceae family was enriched during baseline, emphasizing the effects of previous diet on the microbiota profile.
Studies that have evaluated the impact of low-fiber/high-protein meat-based raw diets in the gut microbiome of healthy dogs have shown an overall decrease in the abundance of Firmicutes, including genera Peptostreptococcus and Faecalibacterium; and in Bacteroides and Prevotella (phylum Bacteroidetes) [22, 23, 49]. Conversely, other bacterial taxa were found to increase in abundance, including Proteobacteria and Fusobacteria (genus Fusobacterium) [22, 23], and two genera from phylum Firmicutes (Lactobacillus and Clostridium) [22, 49].
Although previous studies have identified increased levels of Enterobacteriaceae in dogs fed raw diets, we did not see enrichment of this bacterial group during this dietary intervention [49].
The high-insoluble fiber diet (diet B) used in this study contains 25.5% (dry matter [DM]) insoluble fiber and 1.9% DM soluble fiber; the total dietary fiber is therefore 27.6% DM and crude fiber 16.4% DM. This type of diet is used for conditions such as weight loss, diabetes mellitus, chronic pancreatitis and historically for conditions like colitis. Most standard canine diets fed for maintenance in adults contain crude fiber around 1.5–5% DM. Diet B in our study induced an enrichment in bacteria belonging to the Firmicutes and Actinobacteria phyla. However, at family level, Prevotellaceae (belonging to the Bacteroidetes phylum) was also enriched. This agrees with human studies, where it has been found that increased levels of Prevotella are associated with a plant-based diet rich in fiber, simple sugars, and plant-derived compounds, as they harbor genes for cellulose and xylan hydrolysis [38, 50].
Hydrolyzed diets are used frequently in dogs for the treatment of putative food allergies and chronic enteropathy [30, 33, 51]. The main difference between a commercial dry diet designed for healthy dogs and a hydrolyzed diet is that the latter is composed of smaller chains of protein or single amino acids that decreases the probability of an immune response to protein dietary components [30]. The diet used (diet A) is based on hydrolyzed poultry, and although has lower fiber content and higher fat content than diet B, it is similar in overall macronutrient composition to commercial maintenance diets. Evaluation of the effect of the hydrolyzed diet did not show overrepresentation of any member at the phylum and family phylogenetic levels, in comparison with the other two diets. Potentially, dietary impact of hydrolyzed diets on the gut microbiota could be at functional level and not necessarily at taxonomic levels. Although we used PICRUSt to predict community functional capabilities [39], we could not determine if any functional changes had actually occurred. Further studies assessing function (metabolomics), and strains (metagenomics) could help us to elucidate the relevance and the role of these microbiota changes in the gut. It is interesting that these changes were different and independent from the high fiber diet, which suggests a different impact on the microbiota.
Recent studies evaluating the effect of a hydrolyzed diet on the gut microbiome in healthy dogs and in dogs with food-responsive enteropathy showed that the impact of the diet was minimal in the microbial composition as well as in the metabolome [40, 52]. In these studies, dogs were fed with commercial maintenance diets before the introduction of the new diet, whereas in our study the baseline diet was meat-based, which could potentially influence the results. Also, the percentage of fat differs among hydrolyzed diets, with our diet being slighter higher in fat percentage.
We also saw that the magnitude and nature of the changes induced by the high-insoluble fiber and hydrolyzed diets varied according to the diet sequence. The initial bacterial composition, the fact that bacteria form a metabolic network and cross-feed each other and that there is significant heterogeneity within bacterial species in their ability to digest different types of fiber [2, 53] add complexity to the diet-microbiota interaction. In people, particularly in the case of fiber, it has been shown that an individual’s baseline microbiota harbors predictive potential with regards to the effect of dietary constituents on the host [54]. Also, we should take into consideration that the proportion of one macronutrient to the total energy intake inherently influences the contribution from other macronutrients. Thus, the effect of a change in one macronutrient on the fecal microbiota is a result of the combinatory effect of all the macronutrients [55].
In our study, we observed that the ratio of Prevotella to Bacteroides was higher with the hydrolyzed and high-insoluble fiber diets compared to the high-protein diet. In accordance with this, it has been reported that a high fiber diet correlates with a polysaccharide-utilizing microbiota with lower protein fermentation products and fewer Bacteroides and Clostridia [56, 57]. However, when we analyzed the families using the Dirichlet model, we observed that Prevotellaceae was only higher in the high-insoluble fiber diet and only in the ACB sequence, whereas members of the Bacteroidaceae family were higher in the high-protein diet but only during the baseline period.
Finally, assessment of the gut microbiota during the washout period showed that the gut microbiota of dogs did not revert to their original phylogenetic structure after 6 weeks. Previous studies in dogs have reported adaptation periods varying from 10 days to 4 weeks [5, 6, 8, 23, 58]. In our study, although the washout period was longer than previously reported, changes in the composition of the gut microbiota persisted over time. This was evidenced by sequence and diet effects and by differential results in bacterial abundance between baseline and washout periods. These changes could be permanent, or there is a possibility that more time is required to return to baseline levels. The intestinal microbiota is reported to be resistant to most environmental influences, returning rapidly to its pre-treatment state, particularly for short-term interventions, suggesting in our dogs that only diet was responsible [4]. Furthermore, studies have shown that long term changes to dietary habits may be required to achieve permanent changes in the gut community structure [38]. However, this can depend on the magnitude and duration of the change being study [4, 59].
The limitations of this study were the presence of only one breed, age (although they were evenly distributed in both groups), and that potentially the manufacturing process of the commercial diets themselves could have influenced the gut microbiota. The alternate day feeding pattern of diet C could also have influenced microbiome composition independently of the protein/digestibility [60, 61]. Furthermore, day to day variations in microbiota occurs and, in our study, feces were only collected at a set time point [36]. Pooling samples over a collection period of several days may have been more beneficial to average out day-to-day variability but would have added more complexity to the analysis.
In addition, evaluation of microbial composition together with functional analysis (metabolomics, transcriptomics) would offer a better insight into the total effect of diets [53]. Different microbiomes have different potentials for producing certain metabolites, depending on the metabolic capabilities and metabolic interactions within the population. The fact that a bacterium harbors a gene does not imply that the gene is expressed. In the presence of different energy sources, bacteria may express genes to produce one, a group or several of these enzymes, depending on the environmental context [53]. Future studies could combine several approaches to elucidate the influence of the diet-microbiota interaction on host biology.
Conclusion
This study demonstrated that that dietary protein and fiber ratios can impact the gut microbial composition. Alterations on the microbiota structure are dependent on the bacterial composition present at the time of intervention, as results were quite susceptible to study design, evidenced by sequence and diet effects. Further functional studies are required for a better understanding of the ways the dietary-microbiome crosstalk interacts with the host. This will allow, in the future, the implementation of targeted and effective dietary interventions for the alleviation of microbiome-associated diseases.
Methods
Study dogs
Fifty healthy foxhound dogs (all lean body weight, body condition score [BCS] range 3–5/9 Purina body condition score system) were enrolled in the study [62]. All dogs had up to date vaccination status and no signs of gastrointestinal disease or medication within the previous 3 months. All dogs enrolled underwent a full physical examination, complete blood count and biochemical profile. They were dewormed with praziquantel 200 mg, pyrantel 560 mg and oxantel embonate 2180 mg (Paratak™ Plus) on two separate occasions 12 weeks apart prior to commencement of the trial.
The dogs were normally fed a high-protein (all meat/carcass) diet every second day. For the study, the dogs were kept in two groups of 25 each, physically separated during the study. The two groups had access to communal drinking water in their allocated yards, obtained from the same source, and were located close to each other, but had no contact with animals from the other group for the duration of the study. All environmental factors were the same for both groups (shelter, bedding, exercise area etc.).
Group 1 contained 10 males and 13 females with a mean age of 5.3 ± 2.5 (SD). Group 2 contained 16 males and 5 females with a mean age of 3.7 ± 2. Two dogs in group 1 were excluded during the feeding trial period, one due to illness, the other as it refused to eat the trial food. Four dogs were excluded from group 2 during the feeding trial period: three due to antibiotic use and one due to inadequate fecal sample collection at one time point.
Each group was randomly assigned to be fed one of the two experimental diets: diet A (hydrolyzed) (Hill’s® Prescription Diet® z/d® Canine) or diet B (high-insoluble fiber) (Hill’s™ Prescription Diet™ w/d™) diet daily for 6 weeks (Phase 1). Following this, there was a washout phase of 6 weeks when dogs returned to their normal high-protein diet (all meat/carcass, raw diet) (diet C) fed alternate days. The groups were then crossed over to receive the alternative diet for 6 weeks (Phase 2). Dogs were fed to maintain body weight once daily and had free access to water (Fig. 1).
Samples
Individual fecal samples were obtained at 4 time points: baseline, after 6 weeks of the first diet (diet A or B), after 6 weeks of washout (on baseline diet) and after 6 weeks on the second diet (crossing over to A or B). A total of 176 samples were collected.
Diet composition and analysis
The main source of protein in diet A was hydrolyzed chicken liver, whereas for diet B the main source was soybean meal. The main source of carbohydrate (CHO) in diet A was corn starch and cellulose and for diet B was soybean meal. Regarding fiber, diet A was mainly composed of powdered cellulose and diet B of soybean meal and dried beet pulp. A detailed list of ingredients of the commercial diets can be found in Additional file 6: Table S2.
The following analysis of diets A and B were obtained directly from the manufacturer (as diet is not the same as currently produced) (Table 1). The content of diet C (horse meat carcass- bones, muscles, ligaments but no organs) was analyzed using online diet composition and published references of horse meat composition for protein, fat, CHO (http://www.foodnutritiontable.com/nutritions/nutrient/?id=132. Page accessed April 2020) [46, 47].
Table 1.
Nutrient composition of the different diets
| Diet | Fat % Dry matter (DM) | Fat g/100 kcal ME* | Protein % DM | Protein g/100 kcal ME* | Crude fiber % DM | Crude fiber g/100 kcal ME* | CHO (NFE) % DM | CHO (NFE) g/100 kcal ME* | Ash % DM | Kcal/100 g |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 14.4 | 4 | 19 | 5.3 | 2.9 | 0.8 | 58.4 | 16.2 | 5.3 | 360 |
| B | 8.7 | 2.91 | 19.2 | 6.4 | 16.4 | 5.5 | 50.8 | 16.99 | 5.0 | 299 |
| C | 6.63 | 6.14 | 21.1 | 19.54 | 0 | 0 | 0 | 0 | 0.5 | 108 |
A: Hydrolyzed, B: High-insoluble fiber, and C: Horse carcass.*As fed. ME Metabolizable energy, DM Dry matter, NFE Nitrogen-free extract
Fecal DNA extraction
All samples were collected upon voiding without contacting the environment (to avoid transfer of genetic material) or via rectal collection. Samples were refrigerated at 4 °C until transport to the laboratory, which was completed within 48 h of sample collection. Samples were then frozen and stored at − 80 °C until processing.
Fecal DNA was extracted using the Power soil DNA isolation kit (MoBio® laboratories Catalog No. 12888–100); 250 mg of feces were processed using the protocol for DNA isolation, detailed in the manufacturer’s instructions, with some modifications. Briefly, the fecal pellet was added to a glass bead tube (0.1 mm) and 750 μL of bead solution and 60 μL of C1 solution were added. Then, samples were incubated at 94 °C for 10 min. Afterwards, tubes were placed in the PowerLyzer® 24 and were run at 3000 rpm for 45 s. Subsequent steps were done as indicated by the manufacturer. Extracted DNA was eluted from the spin column in 100 μL of C6 solution from Mobio® (10 mM tris-Cl pH 8.0–8.5). Extracted DNA was quantified and checked for purity, based on UV absorption ratios 260:280 nm and 260:230 nm, on a ND1000 spectrometer (NanoDrop™ 2000/2000c). Samples with highly aberrant absorption ratios were re-extracted.
Bacterial 16S rRNA gene analysis
Illumina sequencing of the V4 region of the bacterial 16S rRNA genes was performed using primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) to 806R (5′- GGACTACVSGGGTATCTAAT-3′). Raw data was analyzed using the open-source software package Quantitative Insights into Microbial Ecology (QIIME) [63]. Version 1 (QIIME1, release 1.9.0). The sequence data was demultiplexed, and then quality filtered using the default settings for QIIME. Chimeras were detected and filtered from the reads using USEARCH [64] against the 97% clustered representative sequences from the Greengenes v 13.8 database [65]. The remaining sequences were clustered into Operational Taxonomic Units (OTUs) by using an open reference approach in QIIME [63].
From 176 samples, a total of 11.650.924 high-quality sequences were obtained, with the number of reads ranging from 12.391 to 165.430 per sample (median 60.448; mean 65.824.429; standard deviation (SD) 29.494.371). Samples were rarefied at 12.390 sequences per sample for even depth of analysis.
Rarefaction plots were used to visualize adequacy of depth in the sequencing data. Measurements of Alpha (α) – diversity and beta (ß)-diversity were done using QIIME1 and Phyloseq package (version1.18.1) [66]. Beta-diversity was assessed using the Bray–Curtis dissimilarity metrics.
Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) [39] was used to predict functional gene content based on 16S rRNA gene data present in the Greengenes database and the KEGG database, using the “predict_metagenomes.py” command in PICRUSt (v1.0.0) (http://picrust.github.io/picrust/).
Statistical analyses
Shannon index (alpha diversity) was defined as the response in a linear mixed model, which included a subject-level random intercept. Fixed effects were the diet, sequence, and interaction of diet and sequence. The model was defined using the ‘lme4’ package in R [67]. Informativeness of the model was assessed using the marginal and conditional coefficients of determination as implemented in the ‘muMin’ package [35, 68].
The same model was used for predictive functional analysis. The L2 level was chosen, and the pathway was defined as the response in the linear mixed model which included a subject-level random intercept. The responses were log-transformed for the analysis. The package ‘emmeans’ from R, was used for post-hoc comparisons among diets and sequences and for estimating marginal means and their 95% confidence intervals [69].
Associations between the diet, and sequence, and the relative abundance of phyla and families were assessed using a hierarchical Dirichlet regression model with the logit link function [70]. The response variables were the proportional abundances of 20 families, where Bifidobacteriaceae was the reference level, or the proportional abundances of 5 phyla, where Actinobacteria was the reference level. The sum-to-one compositional constraint in the family abundances was handled by the Dirichlet response distribution. A handful of zeros in the original abundance data, disallowed in the Dirichlet distribution, were arbitrarily adjusted, and an ‘OTHER’ category was generated to capture the proportion remaining (satisfying the sum-to-one constraint). Between-dog variability in the intercept for each bacterial family was estimated to accommodate the repeated-measures structure. The model was implemented in R [71] using the ‘brms’ package [72]. The MCMC (Markov chain Monte Carlo) sampling used 4 chains of 10,000 iterations. Chain convergence was assessed visually and by the potential scale reduction statistic R^. Priors for the regression coefficients were set as N(0,5), intended to be minimally informative. Due to interpretational difficulty associated with the interdependence of the parameter estimates across families and phyla, the final model was assessed using the posterior predicted abundances across groups and their 89% prediction intervals (considering the residual variation) (‘predicted’ interval plots) and are represented in Fig. 4 (phylum) and 5 (family). The posterior estimated population mean of relative abundances with 89% credible intervals (‘fitted’ interval plots) can be found in Additional file 7: Fig. 5 (phylum) and Additional file 8: Fig. 6 (family). Values of the ‘predicted’ and ‘fitted’ interval plots at phylum and family level are also reported (See Additional file 9: Table S3 [phylum] and Additional file 10: Table S4 [family]).
Supplementary Information
Additional file 1: Figure S1. Relative abundance of bacteria before (high-protein) and after the introduction of the new diet. A: Hydrolyzed diet (diet A) B: High-insoluble fiber diet (diet B). Top 5 most abundant phyla. a: Baseline [high-protein] (diet C) b: Washout [high-protein] (diet C). Median with range.
Additional file 2: Table S1. Estimates of the Linear mixed model for Shannon Index.
Additional file 3: Figure S2. A: PCoA of Bray-Curtis dissimilarity index on diet sequence ACB and distributions of samples along the PC1 by diet. The percentage of variation explained by the principal coordinates (PC1 and PC2) is indicated on the axes. B: Bray-Curtis distance boxplots of the differences in relative abundance between the baseline and the post-treatment sample from the same dog, in diet sequence ACB. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 4: Figure S3. A: PCoA of Bray-Curtis dissimilarity index on diet sequence BCA and distributions of samples along the PC1 by diet. The percentage of variation explained by the principal coordinates (PC1 and PC2) is indicated on the axes. B: Bray-Curtis distance boxplots of the differences in relative abundance between the baseline and the post-treatment sample from the same dog, in diet sequence BCA Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 5: Figure S4. A: Prevotella and B: Bacteroides relative abundances as a function of the diet. C: Ratios between the two genera in the different categories of diet.
Additional file 6: Table S2. List of ingredients commercial diets.
Additional file 7: Figure S5. The posterior estimated population mean of relative abundances at phylum level in diet sequence ACB and BCA. Points are the posterior population mean. The bars are the 89% credible intervals. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 8: Figure S6. The posterior estimated population mean of relative abundances at family level in diet sequence ACB and BCA. Top of the 20 most abundant families. Points are the posterior population mean. The bars are the 89% credible intervals. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 9: Table S3. Hierarchical Dirichlet regression model values of the (A) ‘predicted’ and (B) ‘fitted’ interval plots at phylum level.
Additional file 10: Table S4. Hierarchical Dirichlet regression model values of the (A) ‘predicted’ and (B) ‘fitted’ interval plots at family level.
Acknowledgements
The authors acknowledge the support of Leilani Santos and Louise Baker during the experiments. We acknowledge the advice of Dr. Raphael Trouve regarding the statistical model.
Abbreviations
- BCS
Body condition score
- CHO
Carbohydrate
- DM
Dry matter
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MW
Molecular weight
- MCMC
Markov chain Monte Carlo
- NCBI
National center for biotechnology information
- NFE
Nitrogen-free extract
- NHMRC
National Health and Medical Research Council
- OTU
Operational taxonomic unit
- PC1
Principal-coordinate 1
- PC2
Principal-coordinate 2
- PCoA
Principal-coordinate analysis
- PERMANOVA
Permutational multivariate analysis of variance
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- QIIME
Quantitative Insights into Microbial Ecology
- SD
Standard deviation
- SRA
Sequence Read Archive
Authors’ contributions
Conceived and designed the experiment: CM. Performed the experiments: AP, LM. Microbial data analysis: RP, LM, AW, CM, JS. Statistical analysis: LM, AW. Drafting the paper: AP, LM, AW, CM. Paper revisions and final approval: AP, LM, RP, AW, JS, CM. The author(s) read and approved the final manuscript.
Funding
The authors received funding for this work from Hills Pet Nutrition and the University of Melbourne residency grant program.
Availability of data and materials
Sequence data generated during this study are available through National center for biotechnology information (NCBI)‘s Sequence Read Archive (SRA) under the BioProject number PRJNA641482. Codes for analysis of the 16S rRNA gene sequencing data, for generation of the figures and for the statistics can be found in the following Github repository: https://github.com/Lina-Maria/Cross-over-diet-study. All other data is included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
All animal procedures were done in accordance with the Animal Ethics committee of University of Melbourne (Animal Ethics Committee approval AEC # 1312931.1), using National Health and Medical Research Council (NHMRC) guidelines. Owner gave written consent and was able to withdraw animals from the trial at any point.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lina María Martínez-López, Email: lina.martinez@unimelb.edu.au.
Amy Pepper, Email: amyepepper@gmail.com.
Rachel Pilla, Email: rpilla@cvm.tamu.edu.
Andrew P. Woodward, Email: andrew.woodward@unimelb.edu.au
Jan S. Suchodolski, Email: jsuchodolski@cvm.tamu.edu
Caroline Mansfield, Email: cmans@unimelb.edu.au.
References
- 1.Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16(4):7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills S, et al. Precision nutrition and the microbiome, part i: current state of the science. Nutrients. 2019;11(4):923. doi: 10.3390/nu11040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Middelbos IS, Vester Boler BM, Qu A, White BA, Swanson KS, Fahey GC. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One. 2010;5(3):e9768. doi: 10.1371/journal.pone.0009768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panasevich MR, Kerr KR, Dilger RN, Fahey GC, Jr, Guérin-Deremaux L, Lynch GL, Wils D, Suchodolski JS, Steer JM, Dowd SE, Swanson KS. Modulation of the faecal microbiome of healthy adult dogs by inclusion of potato fibre in the diet. Br J Nutr. 2015;113(1):125–133. doi: 10.1017/S0007114514003274. [DOI] [PubMed] [Google Scholar]
- 7.Panasevich MR, Rossoni Serao MC, de Godoy MRC, Swanson KS, Guérin-Deremaux L, Lynch GL, Wils D, Fahey GC, Jr, Dilger RN. Potato fiber as a dietary fiber source in dog foods. J Anim Sci. 2013;91(11):5344–5352. doi: 10.2527/jas.2013-6842. [DOI] [PubMed] [Google Scholar]
- 8.Beloshapka AN, Dowd SE, Suchodolski JS, Steiner JM, Duclos L, Swanson KS. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol Ecol. 2013;84(3):532–541. doi: 10.1111/1574-6941.12081. [DOI] [PubMed] [Google Scholar]
- 9.Hang I, Rinttila T, Zentek J, Kettunen A, Alaja S, Apajalahti J, Harmoinen J, de Vos WM, Spillmann T. Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Vet Res. 2012;8(1):90. doi: 10.1186/1746-6148-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilla R, Suchodolski JS. The role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front Vet Sci. 2020;6:498. doi: 10.3389/fvets.2019.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhoutte T, Huys G, Brandt E, Fahey GC, Jr, Swings J. Molecular monitoring and characterization of the faecal microbiota of healthy dogs during fructan supplementation. FEMS Microbiol Lett. 2005;249(1):65–71. doi: 10.1016/j.femsle.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol Ecol. 2010;71(2):304–312. doi: 10.1111/j.1574-6941.2009.00812.x. [DOI] [PubMed] [Google Scholar]
- 13.Simpson JM, Martineau B, Jones WE, Ballam JM, Mackie RI. Characterization of fecal bacterial populations in canines: effects of age, breed and dietary fiber. Microb Ecol. 2002;44(2):186–197. doi: 10.1007/s00248-002-0001-z. [DOI] [PubMed] [Google Scholar]
- 14.Barry KA, Hernot DC, Middelbos IS, Francis C, Dunsford B, Swanson KS, Fahey GC. Low-level fructan supplementation of dogs enhances nutrient digestion and modifies stool metabolite concentrations, but does not alter fecal microbiota populations. J Anim Sci. 2009;87(10):3244–3252. doi: 10.2527/jas.2008-1659. [DOI] [PubMed] [Google Scholar]
- 15.Biagi G, Cipollini I, Grandi M, Zaghini G. Influence of some potential prebiotics and fibre-rich foodstuffs on composition and activity of canine intestinal microbiota. Anim Feed Sci Technol. 2010;159(1):50–58. doi: 10.1016/j.anifeedsci.2010.04.012. [DOI] [Google Scholar]
- 16.Graham PA, et al. Influence of a high fibre diet on glycaemic control and quality of life in dogs with diabetes mellitus. J Small Anim Pract. 2002;43(2):67–73. doi: 10.1111/j.1748-5827.2002.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 17.Herstad KMV, Gajardo K, Bakke AM, Moe L, Ludvigsen J, Rudi K, Rud I, Sekelja M, Skancke E. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet Res. 2017;13(1):147. doi: 10.1186/s12917-017-1073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hang I, Heilmann RM, Grützner N, Suchodolski JS, Steiner JM, Atroshi F, Sankari S, Kettunen A, de Vos WM, Zentek J, Spillmann T. Impact of diets with a high content of greaves-meal protein or carbohydrates on faecal characteristics, volatile fatty acids and faecal calprotectin concentrations in healthy dogs. BMC Vet Res. 2013;9(1):201. doi: 10.1186/1746-6148-9-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bermingham EN, Maclean P, Thomas DG, Cave NJ, Young W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ. 2017;5:e3019. doi: 10.7717/peerj.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coelho LP, Kultima JR, Costea PI, Fournier C, Pan Y, Czarnecki-Maulden G, Hayward MR, Forslund SK, Schmidt TSB, Descombes P, Jackson JR, Li Q, Bork P. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome. 2018;6(1):72. doi: 10.1186/s40168-018-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, et al. Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. mBio. 2017;8(1):e01703–16. 10.1128/mBio.01703-16. [DOI] [PMC free article] [PubMed]
- 22.Kim J, An JU, Kim W, Lee S, Cho S. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 2017;9(1):68. doi: 10.1186/s13099-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandri M, Dal Monego S, Conte G, Sgorlon S, Stefanon B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet Res. 2017;13(1):65. doi: 10.1186/s12917-017-0981-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9(10):599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 25.Minamoto Y, Otoni CC, Steelman SM, Büyükleblebici O, Steiner JM, Jergens AE, Suchodolski JS. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. 2015;6(1):33–47. doi: 10.1080/19490976.2014.997612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.AlShawaqfeh MK, et al. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol Ecol. 2017;93(11). 10.1093/femsec/fix136. [DOI] [PubMed]
- 27.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. 2012;7(12):e51907. doi: 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honneffer JB, Minamoto Y, Suchodolski JS. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J Gastroenterol. 2014;20(44):16489–16497. doi: 10.3748/wjg.v20.i44.16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson KW, Jergens AE. Pitfalls and progress in the diagnosis and management of canine inflammatory bowel disease. Vet Clin North Am Small Anim Pract. 2011;41(2):381–398. doi: 10.1016/j.cvsm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Cave NJ. Hydrolyzed protein diets for dogs and cats. Vet Clin North Am Small Anim Pract. 2006;36(6):1251–1268. doi: 10.1016/j.cvsm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Simpson JW. Diet and large intestinal disease in dogs and cats. J Nutr. 1998;128(12 Suppl):2717s–2722s. doi: 10.1093/jn/128.12.2717S. [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Martins R, Sullivan MC, Friedman ES, Misic AM, el-Fahmawi A, de Martinis ECP, O’Brien K, Chen Y, Bradley C, Zhang G, Berry ASF, Hunter CA, Baldassano RN, Rondeau MP, Beiting DP. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7(1):126. doi: 10.1186/s40168-019-0740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandigers PJ, et al. A randomized, open-label, positively-controlled field trial of a hydrolyzed protein diet in dogs with chronic small bowel enteropathy. J Vet Intern Med. 2010;24(6):1350–1357. doi: 10.1111/j.1939-1676.2010.0632.x. [DOI] [PubMed] [Google Scholar]
- 34.Suchodolski JS. Intestinal microbiota of dogs and cats: a bigger world than we thought. Vet Clin North Am Small Anim Pract. 2011;41(2):261–272. doi: 10.1016/j.cvsm.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagawa S, Johnson PCD, Schielzeth H. The coefficient of determination R(2) and intra-class correlation coefficient from generalized linear mixed-effects models revisited and expanded. J R Soc Interface. 2017;14(134):20170213. 10.1098/rsif.2017.0213. [DOI] [PMC free article] [PubMed]
- 36.Garcia-Mazcorro JF, Dowd SE, Poulsen J, Steiner JM, Suchodolski JS. Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiologyopen. 2012;1(3):340–347. doi: 10.1002/mbo3.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hooda S, Boler BMV, Serao MCR, Brulc JM, Staeger MA, Boileau TW, Dowd SE, Fahey GC, Jr, Swanson KS. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr. 2012;142(7):1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 38.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bresciani F, Minamoto Y, Suchodolski JS, Galiazzo G, Vecchiato CG, Pinna C, Biagi G, Pietra M. Effect of an extruded animal protein-free diet on fecal microbiota of dogs with food-responsive enteropathy. J Vet Intern Med. 2018;32(6):1903–1910. doi: 10.1111/jvim.15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suchodolski JS, Camacho J, Steiner JM. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol. 2008;66(3):567–578. doi: 10.1111/j.1574-6941.2008.00521.x. [DOI] [PubMed] [Google Scholar]
- 42.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander DD, Cushing CA. Red meat and colorectal cancer: a critical summary of prospective epidemiologic studies. Obes Rev. 2011;12(5):e472–e493. doi: 10.1111/j.1467-789X.2010.00785.x. [DOI] [PubMed] [Google Scholar]
- 44.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown RG. Digestibility of pet foods. Can Vet J. 1987;28(6):314–315. [PMC free article] [PubMed] [Google Scholar]
- 46.Badiani A, Nanni N, Gatta PP, Tolomelli B, Manfredini M. Nutrient profile of horsemeat. J Food Compos Anal. 1997;10(3):254–269. doi: 10.1006/jfca.1997.0540. [DOI] [Google Scholar]
- 47.Lorenzo JM, Sarriés MV, Tateo A, Polidori P, Franco D, Lanza M. Carcass characteristics, meat quality and nutritional value of horsemeat: a review. Meat Sci. 2014;96(4):1478–1488. doi: 10.1016/j.meatsci.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Diether NE, Willing BP. Microbial fermentation of dietary protein: an important factor in diet−Microbe−Host interaction. Microorganisms. 2019;7(1):19. doi: 10.3390/microorganisms7010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt M, Unterer S, Suchodolski JS, Honneffer JB, Guard BC, Lidbury JA, Steiner JM, Fritz J, Kölle P. The fecal microbiome and metabolome differs between dogs fed bones and raw food (BARF) diets and dogs fed commercial diets. PLoS One. 2018;13(8):e0201279. doi: 10.1371/journal.pone.0201279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marks SL, Laflamme DP, McAloose D. Dietary trial using a commercial hypoallergenic diet containing hydrolyzed protein for dogs with inflammatory bowel disease. Vet Ther. 2002;3(2):109–118. [PubMed] [Google Scholar]
- 52.Pilla R, et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J Vet Intern Med. 2020;34(5):1853–66. [DOI] [PMC free article] [PubMed]
- 53.Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. 2019;17(12):742–753. doi: 10.1038/s41579-019-0256-8. [DOI] [PubMed] [Google Scholar]
- 54.Mills S, et al. Precision nutrition and the microbiome part ii: potential opportunities and pathways to commercialisation. Nutrients. 2019;11(7):1468. 10.3390/nu11071468. [DOI] [PMC free article] [PubMed]
- 55.Hewson-Hughes AK, Hewson-Hughes VL, Colyer A, Miller AT, McGrane SJ, Hall SR, Butterwick RF, Simpson SJ, Raubenheimer D. Geometric analysis of macronutrient selection in breeds of the domestic dog, Canis lupus familiaris. Behav Ecol. 2013;24(1):293–304. doi: 10.1093/beheco/ars168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frame LA, Costa E, Jackson SA. Current explorations of nutrition and the gut microbiome: a comprehensive evaluation of the review literature. Nutr Rev. 2020;78(10):798–812. doi: 10.1093/nutrit/nuz106. [DOI] [PubMed] [Google Scholar]
- 57.Christodoulides S, Dimidi E, Fragkos KC, Farmer AD, Whelan K, Scott SM. Systematic review with meta-analysis: effect of fibre supplementation on chronic idiopathic constipation in adults. Aliment Pharmacol Ther. 2016;44(2):103–116. doi: 10.1111/apt.13662. [DOI] [PubMed] [Google Scholar]
- 58.Kerr KR, Forster G, Dowd SE, Ryan EP, Swanson KS. Effects of dietary cooked navy bean on the fecal microbiome of healthy companion dogs. PLoS One. 2013;8(9):e74998. doi: 10.1371/journal.pone.0074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasiraj AC, et al. The effects of feeding and withholding food on the canine small intestinal microbiota. FEMS Microbiol Ecol. 2016;92(6):fiw085. doi: 10.1093/femsec/fiw085. [DOI] [PubMed] [Google Scholar]
- 61.Kohl KD, Amaya J, Passement CA, Dearing MD, McCue MD. Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol Ecol. 2014;90(3):883–894. doi: 10.1111/1574-6941.12442. [DOI] [PubMed] [Google Scholar]
- 62.Laflamme D. Development and validation of a body condition score system for dogs.: a clinical tool. Canine Pract. 1997;22:10–15. [Google Scholar]
- 63.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 65.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bates D, et al. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. 10.18637/jss.v067.i01.
- 68.Bartoń K. Package ‘MuMIn’:Multi-Model inference. 2020. [Google Scholar]
- 69.Lenth R, et al. Package ‘emmeans’: estimated marginal means, aka least-squares means. 2020. [Google Scholar]
- 70.Douma JC, Weedon JT. Analysing continuous proportions in ecology and evolution: a practical introduction to beta and Dirichlet regression. Methods Ecol Evol. 2019;10(9):1412–1430. doi: 10.1111/2041-210X.13234. [DOI] [Google Scholar]
- 71.Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 72.Bürkner P-C.brms: An R Package for Bayesian Multilevel Models Using Stan. J Stat Softw. 2017;80(1):1–28.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Relative abundance of bacteria before (high-protein) and after the introduction of the new diet. A: Hydrolyzed diet (diet A) B: High-insoluble fiber diet (diet B). Top 5 most abundant phyla. a: Baseline [high-protein] (diet C) b: Washout [high-protein] (diet C). Median with range.
Additional file 2: Table S1. Estimates of the Linear mixed model for Shannon Index.
Additional file 3: Figure S2. A: PCoA of Bray-Curtis dissimilarity index on diet sequence ACB and distributions of samples along the PC1 by diet. The percentage of variation explained by the principal coordinates (PC1 and PC2) is indicated on the axes. B: Bray-Curtis distance boxplots of the differences in relative abundance between the baseline and the post-treatment sample from the same dog, in diet sequence ACB. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 4: Figure S3. A: PCoA of Bray-Curtis dissimilarity index on diet sequence BCA and distributions of samples along the PC1 by diet. The percentage of variation explained by the principal coordinates (PC1 and PC2) is indicated on the axes. B: Bray-Curtis distance boxplots of the differences in relative abundance between the baseline and the post-treatment sample from the same dog, in diet sequence BCA Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 5: Figure S4. A: Prevotella and B: Bacteroides relative abundances as a function of the diet. C: Ratios between the two genera in the different categories of diet.
Additional file 6: Table S2. List of ingredients commercial diets.
Additional file 7: Figure S5. The posterior estimated population mean of relative abundances at phylum level in diet sequence ACB and BCA. Points are the posterior population mean. The bars are the 89% credible intervals. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 8: Figure S6. The posterior estimated population mean of relative abundances at family level in diet sequence ACB and BCA. Top of the 20 most abundant families. Points are the posterior population mean. The bars are the 89% credible intervals. Inter-subject variation is not included. Baseline [high-protein] (diet C), High-Fiber (diet B), Hydrolyzed (diet A) and Washout [high-protein] (diet C).
Additional file 9: Table S3. Hierarchical Dirichlet regression model values of the (A) ‘predicted’ and (B) ‘fitted’ interval plots at phylum level.
Additional file 10: Table S4. Hierarchical Dirichlet regression model values of the (A) ‘predicted’ and (B) ‘fitted’ interval plots at family level.
Data Availability Statement
Sequence data generated during this study are available through National center for biotechnology information (NCBI)‘s Sequence Read Archive (SRA) under the BioProject number PRJNA641482. Codes for analysis of the 16S rRNA gene sequencing data, for generation of the figures and for the statistics can be found in the following Github repository: https://github.com/Lina-Maria/Cross-over-diet-study. All other data is included in this published article and its supplementary information files.