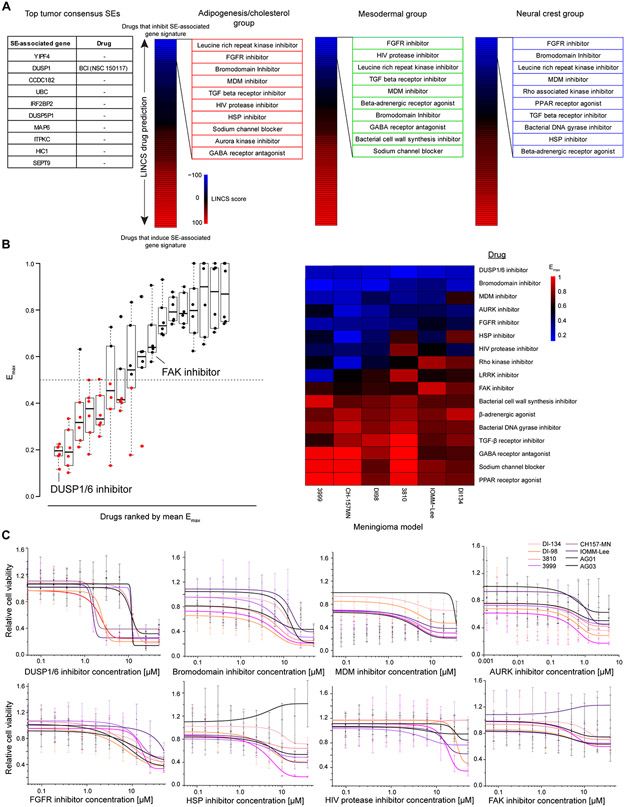
Figure 5. SE-associated genes are critical, druggable targets in vitro.
(A) Selection of drugs based upon SE profiles. Targetable SEs among individual top meningioma super enhancers (SEs) were limited to DUSP1 (left). Signatures were derived using the top 100 subgroup consensus SEs. These gene lists were used as input for the LINCS database which predicts drugs inversely and positively correlated with a gene set. Drugs were ranked from negatively to positively correlated based upon LINCS score. The top 10 compound classes inversely correlated with the SE gene signature from each subgroup were used to derive the drug panel. (B) Results of the screening 17 compounds across 6 meningioma and two arachnoid granulation (AG) models. Maximum drug efficacy (Emax) values are plotted as a boxplot (left), ranked from lowest to highest average Emax . Emax was calculated by comparing all tested drug concentrations to DMSO control and selecting the lowest value. Emax values plotted on a heatmap (right) were ranked from lowest (top, blue) to highest (bottom, red) Emax. Boxplots are represented as the median plus interquartile range. (C) Dose-response curves for each of the 7 compounds with an average Emax<0.5 and the FAK inhibitor. Meningioma models are plotted in shades of red. Normal AG models are in black. Data are represented as mean +/− SD. Sigmoidal curves were fit using a dose-response function.