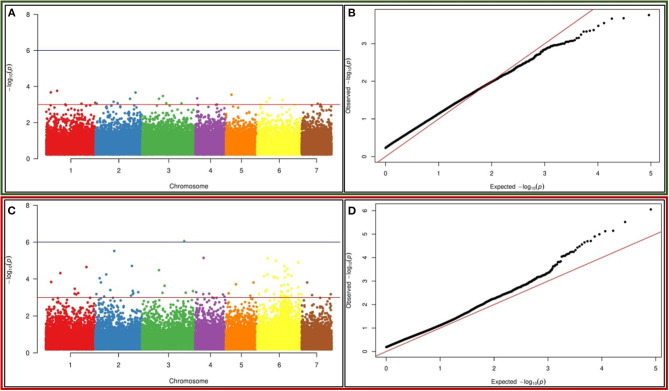
Figure 5.
Genome-wide association studies (GWAS)-based Manhattan plots built in the TASSEL v5.2.64 environment exhibiting significant p-values measured by mixed linear model (MLM) model for (A) DPPH (C) FRAP activity using 67 K SNPs in pearl millet. The x-axis illustrates the relative density of Pennisetum glaucum reference genome-based SNPs physically mapped on seven chromosomes. The y-axis displays the –log10 p-value for the significant association of SNP loci for DPPH the trait. Quantile–quantile plot for (B) DPPH and (D) FRAP activity using the MLM model, built in the TASSEL v5.2.64 environment. The x-axis displayed the expected –log10 p-value and y-axis represented the observed –log10 p-value.