Abstract
We report an automated differentiation model for classifying malignant tumor, fibro-adipose, and stroma in human breast tissues based on polarization-sensitive optical coherence tomography (PS-OCT). A total of 720 PS-OCT images from 72 sites of 41 patients with H&E histology-confirmed diagnoses as the gold standard were employed in this study. The differentiation model is trained by the features extracted from both one standard OCT-based metric (i.e., intensity) and four PS-OCT-based metrics (i.e., phase difference between two channels (PD), phase retardation (PR), local phase retardation (LPR), and degree of polarization uniformity (DOPU)). Further optimized by forward searching and validated by leave-one-site-out-cross-validation (LOSOCV) method, the best feature subset was acquired with the highest overall accuracy of 93.5% for the model. Furthermore, to show the superiority of our differentiation model based on PS-OCT images over standard OCT images, the best model trained by intensity-only features (usually obtained by standard OCT systems) was also obtained with an overall accuracy of 82.9%, demonstrating the significance of the polarization information in breast tissue differentiation. The high performance of our differentiation model suggests the potential of using PS-OCT for intraoperative human breast tissue differentiation during the surgical resection of breast cancer.
1. Introduction
Cancer is the second leading cause of death globally, with an estimated 1.9 million new cancer cases to be diagnosed in the United States in 2021 [1]. Breast, colorectal, lung, cervical, and thyroid cancer are the most common among women. Among all of these, breast cancer is the second leading cause of cancer death in women after lung cancer, accounting for an estimated 284,200 new cases diagnosed and 44,130 deaths in the United States reported in 2021 [1]. The current standard of care for early-stage breast cancer is breast conserving surgery (BCS), which is equivalent to mastectomy in long-term survival rate, preserves more appearance and sensation of the breast, and offers a shorter recovery time [2,3]. Tumor margin evaluation is a crucial issue during BCS in order to avoid margin re-excision and to reduce the risk of local recurrence of the disease. Some evaluative methods have been adopted for intraoperative tumor margin assessment to strive for complete resection of the tumor. Frozen section is time-consuming and provides evaluation results with low accuracy due to difficulties in sectioning adipose tissue [4]. Touch preparation cytology provides faster assessment than frozen section but is associated with sample preparation artifacts [5]. Therefore, there is a significant need for real-time (<1 s) evaluation of the tumor margin during BCS.
Various intraoperative methods based on optical techniques have been reported for breast tumor margin evaluation, including handheld probe-based radiofrequency spectral analysis [6,7], quantitative diffuse reflectance imaging [8,9], confocal mosaicking microscopy [10], point spectroscopy [11,12], and optical coherence tomography (OCT) [13–17]. OCT is a promising method for intraoperative breast tumor margin evaluation because of the high resolution, high acquisition speed for large-area imaging, and millimeter-scale imaging depth [18,19]. Recently with the development of handheld [20] and needle probes [21,22], or utilizing a single fiber [23], portable OCT systems have showed great potential for breast tumor margin evaluation during BCS, especially with the help from advanced artificial intelligence algorithms [24,25]. The criterion for margin evaluation using standard OCT is image contrast and textures, which are dependent on the variations of refractive index and the optical scattering properties of tissues. For breast tissues, it is possible to differentiate between malignant tumor and fibro-adipose tissue types because malignant tumor tissue appears denser and highly scattering than healthy tissue and disrupts the normal low-scattering honeycomb-like architecture of fibro-adipose tissue found in the normal breast [26]. However, normal stroma, which is comprised largely of collagen and connective tissue in the breast, is also dense and highly scattering under standard OCT, making it challenging to differentiate normal stroma from malignant tumor tissue. Recently, OCT-based elastography (OCE) was also applied in breast cancer imaging which showed good performance at detecting normal stroma from malignant tumor tissue by measuring stiffness quantitatively [27]. The promising potential of OCE-based breast tumor margin detection [28–31] was demonstrated and some automated morphological segmentation methods were proposed for breast tissue subtypes differentiation [32–34]. In addition, some works focused on the comparison and combination of OCE and polarization-sensitive OCT [35,36].
In our previous work [37], a bench-top polarization-sensitive OCT (PS-OCT) was demonstrated for detecting and quantifying birefringence contrast between healthy and cancerous tissues based on collagen content, and a portable intraoperative PS-OCT system was used for enhancing the detection and differentiation of invasive ductal carcinoma (IDC) [38]. By using the complementary information provided by standard OCT-based metrics and PS-OCT-based metrics, an overall accuracy of 89.4% was achieved to differentiate fibro-adipose tissue, stroma, and IDC, demonstrating the potential of PS-OCT as an adjunct modality for enhanced intraoperative differentiation of breast cancer. In this paper, we newly report an automated approach for breast tissue differentiation from intraoperative PS-OCT images. Both standard intensity and polarization images were post-processed for further feature extraction. Based on the extracted features, a robust diagnostic model validated by leave-one-site-out-cross-validation (LOSOCV) was built by support vector machine (SVM) to differentiate malignant tumor, fibro-adipose, and stroma tissues using histopathology as the gold standard. The diagnostic model based on features extracted from the combination of one standard OCT-based metric (i.e., intensity) and four PS-OCT-based metrics (i.e., phase difference between two channels (PD), phase retardation (PR), local phase retardation (LPR), and degree of polarization uniformity (DOPU)) lead to a high overall accuracy of 93.5%. The high performance of the classifier suggests that PS-OCT is a promising modality for automated and robust diagnosis of breast cancer, particularly in the intraoperative setting.
2. Materials and methods
2.1. Tissue sample acquisition and preparation
A total of 41 human subjects undergoing surgery for either breast cancer (lumpectomy or mastectomy, all invasive ductal carcinoma (IDC)) or breast reduction surgery (healthy controls, no history of cancer) were recruited for this study. The study protocol was approved by the Institutional Review Boards (IRB) of both Carle Foundation Hospital in Urbana, IL and the University of Illinois at Urbana-Champaign. Tissue specimens from 17 subjects were imaged immediately after surgical resection by the PS-OCT system in the surgical pathology room adjacent to the operating room. The other 24 specimens provided by the Cooperative Human Tissue Network (CHTN) were transported in saline and kept on ice and were imaged within 24 hours of initial surgical resection . After PS-OCT imaging, the imaged sites were marked with surgical ink and the tissue specimens were placed in formalin per standard pathologic protocol. After standard tissue processing and staining with hematoxylin and eosin (H&E) for histological evaluation, diagnoses were made by a board-certified pathologist, and diagnostic results served as the gold standard ground-truth for breast tissue classification.
2.2. PS-OCT imaging
A portable custom-designed PS-OCT system was employed for tissue imaging, which is described in detail in our previous work [38]. Two coupled superluminescent diodes (SLDs) were used in this intraoperative PS-OCT system to cover a spectral range of 1200~1400 nm. The axial resolution and transverse resolution were 5 µm and 8 µm, respectively. Two spectrometers each using a 2048 pixel linescan camera were used for interference signal detection, resulting in a 76 kHz A-scan rate, and the sensitivity of the system was 89 dB. To assess each specimen, one or multiple sites were imaged according to the size of specimen. Each imaged site comprised 512 512 2048 pixels in the X, Y and Z directions, respectively, corresponding to 2.8 2.8 4 mm or 4.2 4.2 4 mm. The scan length in the X and Y directions was determined by the galvo control voltage, which was set to either 4 V or 6 V for different sites, depending on the specimen size.
For each B-scan image, one standard OCT-based metric (i.e., intensity) and four polarization-based metrics (i.e., phase difference between two channels (PD), phase retardation (PR), local phase retardation (LPR), and degree of polarization uniformity (DOPU)) were calculated for the breast tissue classification. For our intraoperative PS-OCT system, the standard OCT intensity I is expressed as the total intensity of the two output channels [39]:
| (1) |
where and are the intensities measured by two spectrometers. The phase difference between the two output channels PD was calculated as
| (2) |
where “*" donates the complex conjugate and “arg" represents the argument.
The PS-OCT enabled PR is represented by
| (3) |
and the local phase retardation LPR is represented by [17]
| (4) |
where , is the matrix logarithm of retardation over a differential depth, is an axial distance, is the Minkowski matrix, and z is a given depth.
To obtain DOPU, the Stokes vectors are first calculated by [40]
| (5) |
where the DOPU is then defined as
| (6) |
with
| (7) |
where i indicates the i-th pixel within a spatial kernel by which DOPU is defined.
2.3. Dataset construction
For multiple sites from the same sample, correlation analyses were conducted in order to ensure data independence and eliminate classification artifacts arising from images with high spatial proximity [38]. The final dataset is summarized in Table 1, noting that 2 subjects contributed to both fibro-adipose and stroma datasets. Therefore, a total of 72 independent sites obtained from 41 patients were finally included in this study. Additionally, we imaged at evenly spaced intervals at each tissue site to get labeled images, but the number of valid labeled images varied significantly among different sites depending on the size and shape of the specimen. To avoid our dataset being partial to subjects/sites with more labeled B-scan images, only 10 images were selected from each site based on the lowest number of labeled frames for all sites. For sites with more than 10 labeled images, we randomly selected 10 images for our dataset.
Table 1. Number of patients, sites, and PS-OCT images included in this study.
| Patients | Sites | PS-OCT images | |
|---|---|---|---|
| Malignant tumor | 10 | 24 | 240 |
| Fibro-Adipose | 15 | 24 | 240 |
| Stroma | 18 | 24 | 240 |
| Total | 41 a | 72 | 720 |
Note that 2 patients contributed to both fibro-adipose and stroma datasets.
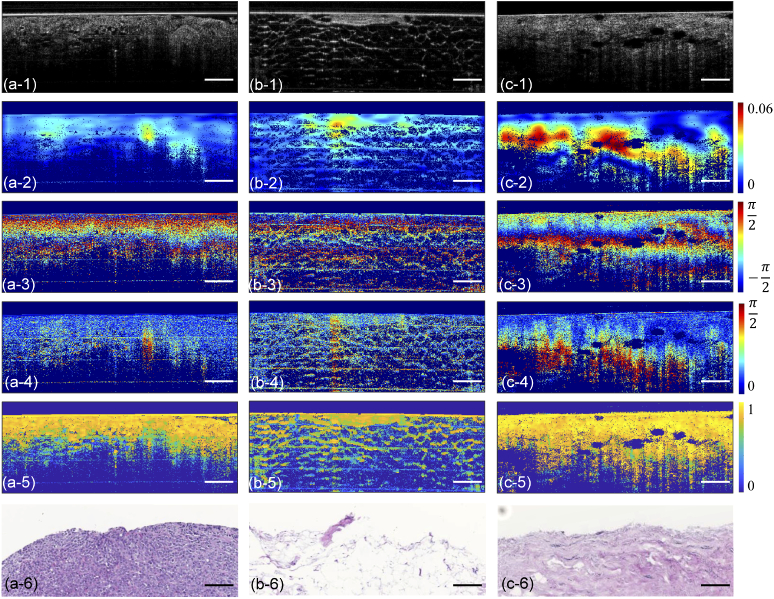
After obtaining raw data from two PS-OCT detection channels, 5 numerical metrics (I, PD, PR, LPR, and DOPU) described in Section 2.2 were calculated in MATLAB for every selected B-scan image. Therefore, 5 metrics matrices of each B-scan image, rather than only the intensity matrix, were used for further feature extraction to take full advantage of the polarization information provided by the PS-OCT system. Figure 1 shows these five metric images along with representative H&E-stained histology results for three tissue types. Thus, the total dataset consisted of 3,600 metric images (720 B-scan images 5 metrics) in this study.
Fig. 1.
Representative structural OCT images (a-1, b-1, c-1), local phase retardation (LPR) images (a-2, b-2, c-2), phase difference (PD) images (a-3, b-3, c-3), phase retardation (PR) images (a-4, b-4, c-4), degree of polarization uniformity (DOPU) images (a-5, b-5, c-5) and H&E-stained histology images (a-6, b-6, c-6) of malignant tumor (a-1 to a-6), fibro-adipose (b-1 to b-6) and stroma (c-1 to c-6) tissue, respectively. Scale bars represent 500 µm. The color scale unit for PD, PR, and LPR is radian, the color scale unit for DOPU is a.u.
2.4. Region of interest (ROI) selection
In OCT systems, signal-to-noise ratio (SNR) decreases in for deeper layers of a sample due to the multiple scattering and overall attenuation in biological tissues. In order to build a model with high reliability, a region-of-interest (ROI) was selected for our dataset to exclude data with low SNR. The upper boundary of the ROI was automatically determined by a greedy algorithm for surface extraction [41]. The lower boundary was determined as a fixed number of pixels below the upper boundary. In this paper, 256 pixels below the upper boundary were chosen, empirically aimed at excluding pixels with low SNR. The number of pixels is constant for every A-scan in this fixed ROI, which is beneficial for further feature extraction.
2.5. Feature extraction
For each selected B-scan image, feature extraction was performed using all 5 of the metrics: I, PD, PR, LPR, and DOPU. Features were extracted from both A-scan line data and B-scan image data to provide local and global information at the same time, where A-scan line data was extracted from B-scan image data in this study. A feature matrix was first built with all the extracted features to cover more image properties, and then a feature subset was selected by forward searching to achieve the highest accuracy and limit overfitting.
2.5.1. A-scan features
Two-dimensional median filters with 8 pixel and 20 pixel kernels in the lateral and axial directions, respectively, were applied to all the B-scan images after ROI selection. The size of the filter was decided heuristically to filter some noise while maintaining and reinforcing the structure of the breast tissue. In this study, the total image dataset contained 3,600 metric images, and each image was comprised of 512 A-scan lines. Thus, it was time-consuming to extract features from all the A-scan lines. While preserving the characteristics of tissue structure, all the image data was down-sampled (retaining every eighth A-scan line) to accelerate A-scan feature extraction. Therefore, there were 64 A-scan lines in each down-sampled B-scan image data.
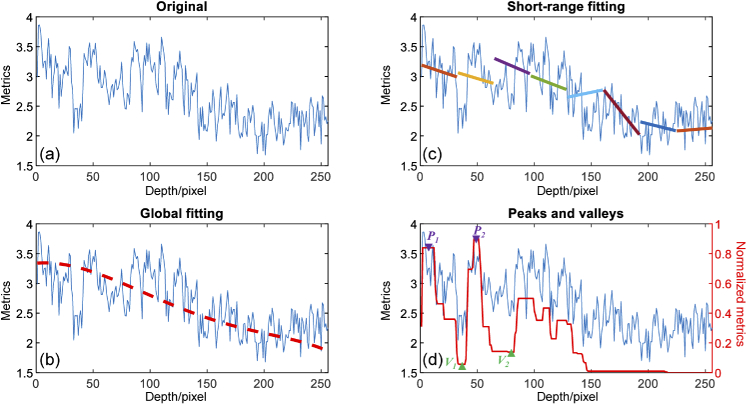
Two groups of features: global features and local features, were calculated for each A-scan line. For global features, polynomial fitting was implemented to all data in the ROI for capturing the changing trend of structures. The polynomial order was primarily determined by tissue type and metric selected. The honeycomb structure of fibro-adipose tissue usually introduced multiple peaks in the depth direction which resulted in a higher order polynomial for better fitting. In addition, in contrast to the intensity metric, some metrics (i.e., PR) became larger for some tissue types at greater depths, which meant that linear regression was not suitable for fitting. In this study, the polynomial fitting order was heuristically set as 5th for all five metrics data. In total, 7 features (6 fitting coefficients and 1 fitting error) were extracted for each metric data as A-scan global features, as shown in Fig. 2(b).
Fig. 2.
A-scan line feature extraction. An original A-scan line data from one image metric is shown in (a). The red dashed line in (b) is the global fitting result after 5th-order polynomial fitting. (c) Short-range linear fitting results in 8 adjacent windows. Peaks and valleys are shown by the red line trace in (d).
Local features consisted of two types: short-range features and peak-valley features. For short-range features, as illustrated in Fig. 2(c), A-scan line data in the ROI was split into 8 consecutive short-range subsets, with 32 pixels in each. Linear fitting was performed in each subset, providing 3 features (slope, intercept, and fitting error) carrying local tissue structure. Five statistical operations: mean, standard deviation (STD), mode, maximum, and minimal were performed among all 8 short-range subsets. Peak-valley features were previously proposed for identifying oral malignancy in the hamster cheek pouch [42]. This type of feature also had the potential to differentiate fibro-adipose from other tissue types due to the regular adipocyte boundaries. By performing morphological closing and opening by flat structuring elements of size 5 and 2 pixels, respectively, A-scan lines were filtered and then normalized. H-maxima transformation was performed for the suppression of those spurious peaks with normalized values less than 0.1. Local maxima and minima were defined as “peak" and “valley", as shown in Fig. 2(d). Seven features were calculated from all the detected peaks and valleys: (1) , (2) , (3) , (4) : A-scan data value at , (5) : A-scan data value at , (6) : A-scan data value at , (7) :A-scan data value at , where and represented i-th peak and valley, respectively.
For all the extracted A-scan features, an intermediate feature matrix was built [43], and four statistics (mean, STD, minimum, and maximum) were calculated among all A-scan lines in the down-sampled B-scan images. In the proposed model, we extracted a total of 116 A-scan features. ((7 global features + 15 short-range features + 7 peak-valley features) 4 statistics).
2.5.2. Texture features
Representing the local variations in one small region of an image, texture is a powerful quantify metric for image analysis. Here, a statistical texture analysis technique based on a gray-level co-occurrence matrix (GLCM) method was implemented for texture features extraction [44]. For L uniform gray levels, GLCM is based on the estimation of the second order joint conditional probability density function, , which means the probability of a pixel with a gray-level value i being d pixels away from a pixel with a gray-level value j in the direction. In our study, we first scaled image data to eight uniform levels, then let d = 1, 2, 4, and 6, and = 0, 45, and 90 deg. Therefore, for a given distance d, an 8 8 matrix, was calculated from for different . For each combination of d and , two features, energy and entropy, were calculated as per Eq. (8) and Eq. (9) [44],
| (8) |
| (9) |
where is the th element of the GLCM for distance d, contributing to a total of 24 features for 3 directions with 4 different distances.
2.5.3. Morphological features
Morphological analysis of OCT images has been reported in the discrimination of freshly-excised specimens of gastrointestinal tissue [45]. Region segmentation was first achieved by a k-means method [46], which partitioned the observation of the image data into k clusters. Typically, three steps were involved in the k-means method. An initial k cluster centroid was first chosen, followed by computation of point-to-cluster distances of all observations, and moving each observation to the closest cluster, finally, the average of the observations in each cluster was recalculated to obtain new k centroid locations. This process was repeated until cluster assignments did not change. In this study, all 5 metrics data for one B-scan were segmented into two to four clusters, which meant that the k-means method was performed three times and in total, 9 regions were obtained. Six features were then extracted per region, as follows:
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
The term is the specific metric value of a pixel within the region and N is the number of pixels of the region. Accordingly, the total number of morphological features is 54 (9 regions times 6 features) for each metric image data.
2.5.4. Feature set combination
After the feature extraction mentioned above, a total of 970 features were extracted for further classifier training and cross-validating, summarized in Table 2.
Table 2. Features list for each image metric data in the classifier.
| Group | Method | Features | Number | |
|---|---|---|---|---|
| A-scan | Global | 5th-order polynomial fitting | fitting coefficients fitting error |
7 |
| Local | Linear fitting for each 32-pixel Α-scan subset (mean, STD, mode, maximum, and minimum) | slope intercept fitting error |
15 | |
| Peak-valley searching | 7 | |||
| Texture | Gray-level co-occurrence matrix (GLCM): 3 directions, 4 distances |
energy entropy |
24 | |
| Morphological | k-means methods: 9 regions | mean normalized mean absolute deviation STD skewness kurtosis |
54 | |
| Total | (7+15+7)×4+24+54=194 features/metric × 5 metrics = 970 features | |||
2.5.5. Feature set fine selection
In order to improve the performance of the classifier and the computing efficiency, the best subset of the features was required for further classification and validation. First, features with the same values for B-scan data from all tissue sites were excluded since they provided no useful information for classification. After that, only 754 features remained for the next step. As the initial features were extracted to capture more tissue properties, some of the remaining 754 features may not be independent or carry little information about the image labels. Features with a large dependency may ensure a high accuracy when taken separately, however, the combinations of them may not necessarily lead to a better performance because they do not give independent information. Therefore, the minimal-redundancy-maximal-relevance (mRMR) criterion [47] was applied, giving a candidate list of features which attributed more relevant information to the labels, and fewer correlation with other features. During mRMR-based feature selection, features were first selected based on the maximal relevance (Max-Relevance) criterion, in which the selected features were required, individually, to have the largest mutual information with the labels. It is likely that features selected by Max-Relevance may have rich redundancy because of the large dependency among these features. Therefore, the minimal redundancy (Min-Redundancy) condition was added to select mutually exclusive features. The resulting classifier built on this fine selected feature subset can achieve both high accuracy and faster speed.
2.6. Classification and validation
Supervised learning was employed here for classification, which uses a training set with known labels to predicate the testing set. Support vector machine (SVM) was used here for developing a diagnostic model. As introduced in Section 2.5.5 high-rank features selected by mRMR, were utilized for classifier training. All extracted features varied greatly due to the different sorts of statistical operations. Therefore, all features needed to be normalized to zero mean and unit variance to prevent some features carrying greater weight. By forward searching [48], classification accuracy with different numbers of features for the classifier can be obtained for the final decision on the feature subset. During the forward searching, the evaluation direction was decided by the feature rank after mRMR feature selection. If several features gave the same overall accuracy in one evaluation step, the feature with the highest mRMR rank among these features was selected as the best individual feature for this step because it carried the highest relevance to the labels and the minimal redundancy with other features.
In order to accurately estimate the performance of the classifier, leave-one-tissue-site-out cross-validation (LOSOCV) was used here to avoid potential correlations among one single tissue site. This approach eliminated the dependency of the classification result on training or on the testing set selection and reduced the risk of over-fitting. Based on the site number, 72 iterations for validation were needed in this study. In each iteration, features from one tissue site were selected as the validation set, while features from the remaining 71 sites were employed as the training set. After all the cross validation, accuracy, sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV) were calculated per diagnostic category from all others to evaluate the performance of our classifier.
3. Results and discussion
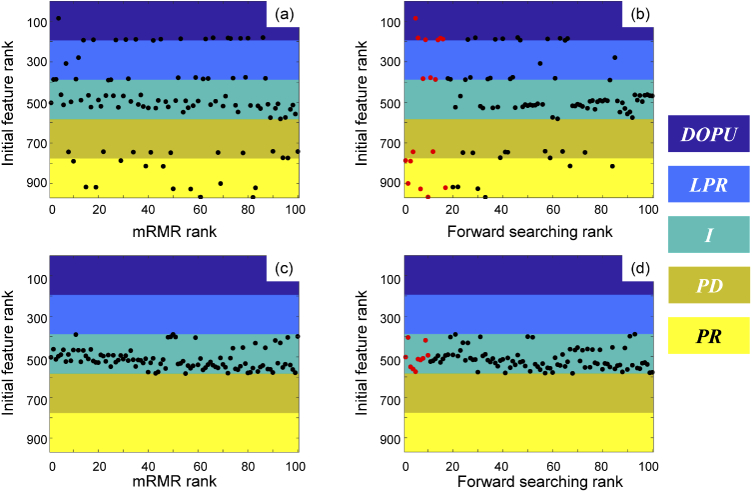
The accuracy of the classifier for differentiating breast tissue mainly depends on the number of features in the subset selected by mRMR criterion. The top 100 high-rank features (47 are from intensity, 11 are from PD, 12 are from PR, 14 are from LPR, and 16 are from DOPU), as illustrated in Fig. 3(a), were first selected based on consideration of the required computing time. Validated by LOSOCV, the overall accuracy for this 100-features-based classifier is 87.2%.
Fig. 3.
Results for best feature set selection. For the PS-OCT-based classifier, (a) is the top 100 high-rank feature distribution sorted by mRMR rank, and (b) is the feature distribution after forward searching, where the best 17 features are marked in red. For the intensity-only classifier, (c) is the top 100 high-rank feature distribution sorted by mRMR rank, and (d) is the feature distribution after forward searching, where the best 10 features are marked in red. Colored regions represent features from degree of polarization uniformity (DOPU, purple), local phase retardation (LPR, blue), intensity (I, turquoise), phase difference (PD, dark yellow) and phase retardation (PR, yellow), respectively.
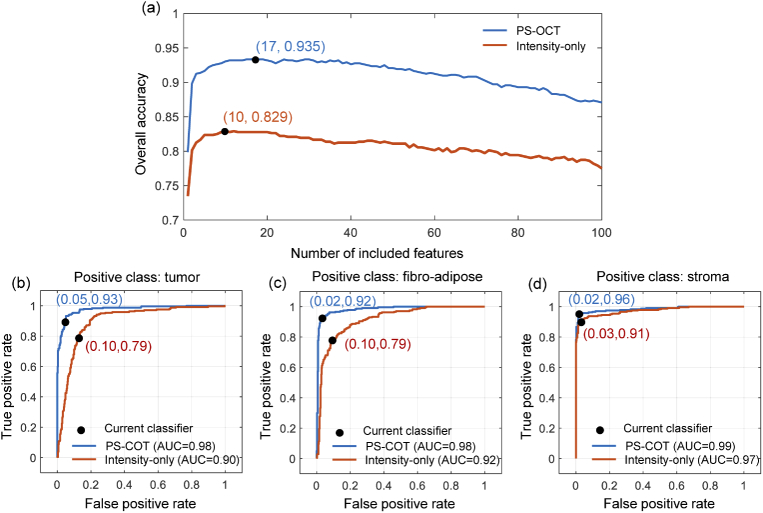
In order to select the best feature subset with the best classification performance, the forward searching algorithm was implemented, and the overall accuracy was calculated as the evaluation metric of classification performance. The feature selected in each forward searching step is shown in Fig. 3(b). The overall accuracy during the forward searching is illustrated as the blue line in Fig. 4(a), where the accuracy reaches a maximum of 93.5% when selecting 17 features (marked in red in Fig. 3(b)) for classifier training. There is no evidence to prove that these 17 features are totally independent, but according to the procedure of mRMR, these features are mutually exclusive features that carried more relevant information to the labels.
Fig. 4.
Prediction performance based on classifier. (a) Overall accuracy during forward searching, using a model trained by 5-metrics-integrated PS-OCT features and intensity-only features, illustrated by the blue and orange lines, respectively. (b)-(d) Receiver operating characteristic (ROC) curves of the 5-metrics-integrated classifier with (b) malignant tumor, (c) fibro-adipose, and (c) stroma as the positive class, respectively.
The 17 features in the final classifier are summarized in Table 3. All of the 17 features were derived from the PS-OCT image metrics (PD: n=2, PR: n=6, LPR: n=3, DOPU: n=6), which also demonstrates that polarization information provided by PS-OCT carried more information about the tissue type. Among the 4 PS-OCT metrics, PR and DOPU contributed more to increase the overall accuracy of the classifier compared to the OCT intensity-only classifier. A total of 13 features in the morphological group made up the largest proportion in the final feature set, mainly because they are more relevant to breast tissue classification, and they occupied a large proportion of the initial feature set (54/194).
Table 3. Number of final selected features in each category.
| Feature group | Metrics | |||||
|---|---|---|---|---|---|---|
| I | PD | PR | LPR | DOPU | ||
| Global | 0 | 0 | 0 | 0 | 0 | |
| A-scan | Short-range | 0 | 0 | 2 | 0 | 0 |
| PV | 0 | 0 | 0 | 0 | 1 | |
| Texture | 0 | 0 | 1 | 0 | 0 | |
| Morphological | 0 | 2 | 3 | 3 | 5 | |
| Subtotal | 0 | 2 | 6 | 3 | 6 | |
| Total | 17 | |||||
The prediction result for this 17-feature-based classifier training is given in Table 4. Among 240 malignant tumor tissue images, 222 were correctly classified, while the remaining 11 were misclassified as adipose and 7 as stroma. For tissue diagnosed as adipose, 221 of 240 were correctly classified as adipose, while 17 were misclassified as malignant tumor and 2 as stroma. Finally, for the stoma group, 230 of 240 were correctly classified as stroma, 9 were misclassified as malignant tumor, and 1 was misclassified as adipose. Based on these results, the obtained sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV), and accuracy for differentiating a breast tissue type from all others are listed in Table 4.
Table 4. Confusion matrix of model trained by integrated features from PS-OCT images with highest overall accuracy by forward searching.
| Histopathology | ||||
|---|---|---|---|---|
| Malignant tumor | Fibro-adipose | Stroma | ||
| Malignant tumor | 222 | 17 | 9 | |
| Predicted | Fibro-adipose | 11 | 221 | 1 |
| Stroma | 7 | 2 | 230 | |
| Sensitivity | 92.5% | 92.1% | 95.8% | |
| Specificity | 94.6% | 97.5% | 98.1% | |
| NPV | 89.5% | 94.9% | 96.2% | |
| PPV | 96.2% | 96.1% | 97.9% | |
| Accuracy | 93.9% | 95.7% | 97.4% | |
To investigate the improvement of classification performance brought by polarization-based information obtained with the use of the PS-OCT system, only intensity data was used for building a simplified classifier for comparison. Feature selection by mRMR, as illustrated in Fig. 3(c), and forward searching were also performed for this intensity-only classifier, whose performance is illustrated by the orange line in Fig. 4(a). Trained by 10 features, as indicated by the red points in Fig. 3(d), the intensity-only classifier reached its best result with an overall accuracy of 82.9%. The receiver operating characteristics (ROC) curves achieved by the 17-feature-based integrated classifier are illustrated by the blue lines in Fig. 4(b)-(d), with malignant tumor (Fig. 4(b)), fibro-adipose (Fig. 4(c)), and stroma (Fig. 4(d)) as the positive classes, and the area-under-curve (AUC) values of 0.98, 0.98, and 0.99, respectively. The ROC curves for the intensity-only classifier are illustrated by the orange lines in Fig. 4(b)-(d), and the AUC values were 0.90, 0.92, and 0.97 for malignant tumor, fibro-adipose, and stroma, respectively. The differentiation model based on the integrated features has higher credibility than intensity-only model. This demonstrated that with the polarization data provided by a PS-OCT system, more information about the tissue type can be obtained. With this extra tissue information, the differentiation model for breast tissue will give a more accurate prediction for an unknown tissue type.
Some limitations of the current differentiation model, and further work for optimization, are to be noted. Although only 17 features were selected for the final model training, initial feature extraction (20 s / PS-OCT image 720 PS-OCT images = 4 hours) and forward searching for the final subset (5 hours) were time consuming (Note the feature extraction and forward searching were operated using MATLAB on a Windows desktop with an Intel Core i7 2.9GHz and 16GB RAM). Additionally, the rank of features determined by mRMR varied considerably for different training datasets, which introduces additional computational time and effort. However, considered from another perspective, this large amount of alternative initial feature set parameters gives more degrees of freedom for the training classifier than those from specific fixed-number feature extraction, which will also improve the prediction accuracy. The time consuming procedure of extracting 970 features and forward searching was only executed during the training of the model. Once the final model was built, for a new unknown breast tissue specimen and image, the tissue type classification will be very quick (<1 s) because only 17 features need to be calculated, highlighting the potential for real-time clinical application in the future. Furthermore, if there is a need to accelerate the testing procedure, we can only select the first several features based on the forward searching result, after trading off the testing time and overall accuracy.
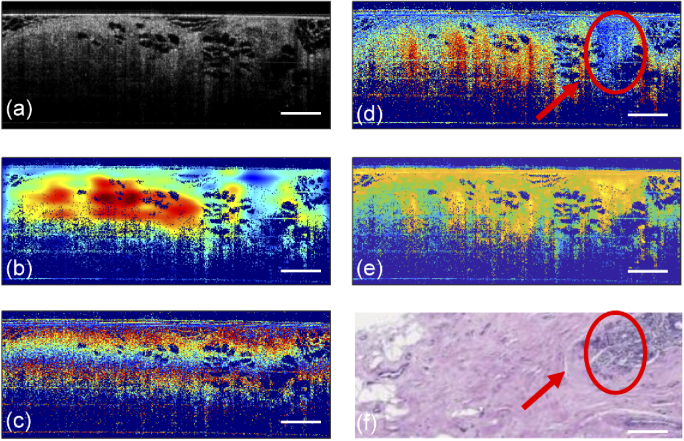
In addition, because the current model is trained through a supervised learning method, the three types of tissues included in the training set were quite homogeneous, with little or no regions within the images being other tissue types. This was intentionally selected to make sure further computation of the five numerical metrics used for feature extraction were corresponding to labels in the classifier training. In clinical settings, the tissues encountered are more likely to be heterogeneous, with a mixture of malignant tumor and normal regions, as shown in Fig. 5. Only a very small region was malignant tumor while most of the image was stroma. From a clinical perspective, the histology result would identify this image as containing malignant tumor, as indicated by the red arrow in Fig. 5(f). However, it was misclassified as stroma because the malignant tumor region was very small relative to the other dominant tissue type, and the current model only performed classification. Image segmentation was not included in our algorithm at the present stage. However, in the near future, we will add a segmentation algorithm to our model to find important sub-ROIs. Further sub-ROI selection can be employed for heterogeneous tissue images, followed by calculating numerical metrics for each sub-ROI. Using the features from those metrics, the predictions for each selected sub-ROI can be obtained. Another alternative method would be to train the differentiation model based on weakly-supervised or unsupervised methods, which is an ongoing project in our group. In weakly-supervised or unsupervised methods, not all training data needs to be labeled, with the potential for margin assessment in clinical applications to be achieved with reliable accuracy.
Fig. 5.
Structural OCT (a), LPR (b), PD (c), PR (d), DOPU (e), and H&E-stained histology (f) images for a heterogeneous tissue region. Region indicated by the red ovals and arrows was identified histologically as malignant tumor.
Finally, all the images in this study were imaged ex-vivo after the tissues were surgically resected from the recruited subjects. Handhold probes for intraoperative OCT imaging of the in-vivo surgical resection bed and loco-regional lymph nodes has been demonstrated [20]. Current efforts are underway for the construction and implementation of a handheld PS-OCT surgical probe that will be integrated with the intraoperative PS-OCT system for in-vivo imaging during malignant tumor resection and for real-time margin assessment with the implementation of our tissue differentiation model.
4. Conclusions
In this work, a differentiation model for breast tissues (malignant tumor, fibro-adipose, stroma) was obtained based on information provided by a lab-built intraoperative PS-OCT system. A total of 72 sites from 41 human subjects were included in training set, with 10 B-scan frames from each site extracted for computing five numerical metrics (I, PD, PR, LPR, DOPU) for each frame. After fine feature subset selection by mRMR and forward searching, the classifier trained by SVM was obtained with an overall accuracy of 93.5% for differentiating malignant tumor, fibro-adipose, and stroma breast tissues. After comparing with a classifier based only on intensity (usually obtained from standard OCT systems), our multiple-metrics-integrated classifier showed better differentiation performance. This demonstrates that with additional polarization information provided by a PS-OCT system, more information about the tissue can be acquired and utilized by multiple numerical metrics, illustrating the potential of PS-OCT for enhanced breast cancer detection. With future integration of a handhold surgical imaging PS-OCT probe, real-time in-vivo margin assessment with machine-learning-based tissue differentiation is anticipated for surgical breast tumor procedures.
Acknowledgments
The authors want to thank the research and surgical nursing staff at Carle Foundation Hospital, Urbana, IL, for assisting with tissue resection, tissue handling, and histopathological evaluation, as well as the human subjects who participated in this research. We also would like to thank the Cooperative Human Tissue Network (CHTN) for providing additional tissue specimens for this study. Additional information can be found at: https://biophotonics.illinois.edu.
Funding
National Institutes of Health10.13039/100000002 (1R01EB023232, IR01CA213149).
Disclosures
S.A.B. is co-founder of Diagnostic Photonics, which is licensing intellectual property from the University of Illinois at Urbana-Champaign related to Interferometric Synthetic Aperture Microscopy for intraoperative imaging of cancer. All other authors declare that there are no conflicts of interest related to this article.
References
- 1.Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., “Cancer statistics, 2021,” CA: A Cancer J. for Clin. 71(1), 7–33 (2021). 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Veronesi U., Cascinelli N., Mariani L., Greco M., Saccozzi R., Luini A., Aguilar M., Marubini E., “Twenty-year follow-up of a randomized study comparing breast-conserving surgery with radical mastectomy for early breast cancer,” N. Engl. J. Med. 347(16), 1227–1232 (2002). 10.1056/NEJMoa020989 [DOI] [PubMed] [Google Scholar]
- 3.Hwang E. S., Lichtensztajn D. Y., Gomez S. L., Fowble B., Clarke C. A., “Survival after lumpectomy and mastectomy for early stage invasive breast cancer: the effect of age and hormone receptor status,” Cancer 119(7), 1402–1411 (2013). 10.1002/cncr.27795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cendan J. C., Coco D., Copeland E. M., “Accuracy of intraoperative frozen-section analysis of breast cancer lumpectomy-bed margins,” J. Am. Coll. Surg. 201(2), 194–198 (2005). 10.1016/j.jamcollsurg.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 5.Valdes E. K., Boolbol S. K., Ali I., Feldman S. M., Cohen J. M., “Intraoperative touch preparation cytology for margin assessment in breast-conservation surgery: does it work for lobular carcinoma?” Ann. Surg. Oncol. 14(10), 2940–2945 (2007). 10.1245/s10434-007-9364-1 [DOI] [PubMed] [Google Scholar]
- 6.Schnabel F., Boolbol S., Gittleman M., Karni T., Tafra L., Feldman S., Police A., Friedman N., Karlan S., Holmes D., Willey S., Carmon M., Fernandez K., Akbari S., Harness J., L. Guerra M. D., Frazier T., Lane K., Simmons R., Allweis T., “A randomized prospective study of lumpectomy margin assessment with use of marginprobe in patients with nonpalpable breast malignancies,” Ann. Surg. Oncol. 21(5), 1589–1595 (2014). 10.1245/s10434-014-3602-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thill M., “Marginprobe: intraoperative margin assessment during breast conserving surgery by using radiofrequency spectroscopy,” Expert Rev. Med. Devices 10(3), 301–315 (2013). 10.1586/erd.13.5 [DOI] [PubMed] [Google Scholar]
- 8.Brown J. Q., Bydlon T. M., Kennedy S. A., Caldwell M. L., Gallagher J. E., Junker M., Wilke L. G., Barry W. T., Geradts J., Ramanujam N., “Optical spectral surveillance of breast tissue landscapes for detection of residual disease in breast tumor margins,” PLoS One 8(7), e69906 (2013). 10.1371/journal.pone.0069906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tromberg B. J., Cerussi A., Shah N., Compton M., “Imaging in breast cancer: diffuse optics in breast cancer: detecting tumors in pre-menopausal women and monitoring neoadjuvant chemotherapy,” Breast Cancer Res. 7(6), 279–285 (2005). 10.1186/bcr1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abeytunge S., Li Y., Larson B., Peterson G., Seltzer E., “Confocal microscopy with strip mosaicing for rapid imaging over large areas of excised tissue,” J. Biomed. Opt. 18(6), 061227 (2013). 10.1117/1.JBO.18.6.061227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller M. D., Vargis E., Mahadevan-Jansen A., de Matos Granja N., Wilson R. H., Mycek M.-A., Kelley M. C., “Development of a spatially offset Raman spectroscopy probe for breast tumor surgical margin evaluation,” J. Biomed. Opt. 16(7), 077006 (2011). 10.1117/1.3600708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer G. M., Zhu C., Breslin T. M., Xu F., Gilchrist K. W., Ramanujam N., “Comparison of multiexcitation fluorescence and diffuse reflectance spectroscopy for the diagnosis of breast cancer,” IEEE Trans. Biomed. Eng. 50(11), 1233–1242 (2003). 10.1109/TBME.2003.818488 [DOI] [PubMed] [Google Scholar]
- 13.Boppart S. A., Luo W., Marks D. L., Singletary K. W., “Optical coherence tomography: feasibility for basic research and image-guided surgery of breast cancer,” Breast Cancer Res. Treat. 84(2), 85–97 (2004). 10.1023/B:BREA.0000018401.13609.54 [DOI] [PubMed] [Google Scholar]
- 14.Nguyen F. T., Zysk A. M., Chaney E. J., Kotynek J. G., Boppart S. A., “Intraoperative evaluation of breast tumor margins with optical coherence tomography,” Cancer Res. 69(22), 8790–8796 (2009). 10.1158/0008-5472.CAN-08-4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zysk A. M., Chen K., Gabrielson E., Tafra L., May Gonzalez E. A., Canner J. K., Schneider E. B., Cittadine A. J., Carney P S., Boppart S. A., Tshuchiya K., Sawyer K., Jacobs L. K., “Intraoperative assessment of final margins with a handheld optical imaging probe during breast-conserving surgery may reduce the reoperation rate: results of a multicenter study,” Ann. Surg. Oncol. 22(10), 3356–3362 (2015). 10.1245/s10434-015-4665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zysk A. M., Boppart S. A., “Computational methods for analysis of human breast tumor tissue in optical coherence tomography images,” J. Biomed. Opt. 11(5), 054015 (2006). 10.1117/1.2358964 [DOI] [PubMed] [Google Scholar]
- 17.Villiger M., Lorenser D., Mclaughlin R. A., Quirk B. C., Kirk R. W., Bouma B. E., Sampson D. D., “Deep tissue volume imaging of birefringence through fibre-optic needle probes for the delineation of breast tumour,” Sci. Rep. 6(1), 28771 (2016). 10.1038/srep28771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha R., Friedlander L. C., Hibshoosh H., Hendon C., Feldman S., Ahn S., Schmidt H., Akens M. K., Fitzmaurice M., Wilson B. C., Mango V. L., “Optical coherence tomography: a novel imaging method for post-lumpectomy breast margin assessment-a multi-reader study,” Academic Radiol. 25(3), 279–287 (2018). 10.1016/j.acra.2017.09.018 [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H., Connolly C., Jaffer S., Oza T., Weltz C. R., Port E. R., Corben A., “Evaluation of surgically excised breast tissue microstructure using wide-field optical coherence tomography,” Breast J. 26(5), 917–923 (2020). 10.1111/tbj.13663 [DOI] [PubMed] [Google Scholar]
- 20.Erickson-Bhatt S. J., Nolan R. M., Shemonski N. D., Adie S. G., Putney J., Darga D., McCormick D. T., Cittadine A. J., Zysk A. M., Marjanovic M., Chaney E. J., Monroy G. L., South F. A., Cradock K. A., Liu Z. G., Sundaram M., Ray P. S., Boppart S. A., “Real-time imaging of the resection bed using a handheld probe to reduce incidence of microscopic positive margins in cancer surgery,” Cancer Res. 75(18), 3706–3712 (2015). 10.1158/0008-5472.CAN-15-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin R., Quirk B., Curatolo A., Kirk R., Scolaro L., Lorenser D., Robbins P., Wood B., Saunders C., Sampson D., “Imaging of breast cancer with optical coherence tomography needle probes: feasibility and initial results,” IEEE J. Sel. Top. Quantum Electron. 18(3), 1184–1191 (2012). 10.1109/JSTQE.2011.2166757 [DOI] [Google Scholar]
- 22.Zysk A. M., Nguyen F. T., Chaney E. J., Kotynek J. G., Boppart S. A., “Clinical feasibility of microscopically-guided breast needle biopsy using a fiber-optic probe with computer-aided detection,” Technol. Cancer Res. Treat. 8(5), 315–321 (2009). 10.1177/153303460900800501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Hubbi B., Liu X., “Single fiber OCT imager for breast tissue classification based on deep learning,” in Optical Fibers and Sensors for Medical Diagnostics and Treatment Applications XX, vol. 11233 (Proc. SPIE, 2020), pp. 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butola A., Prasad D. K., Ahmad A., Dubey V., Qaiser D., Srivastava A., Senthilkumaran P., Ahluwalia B. S., Mehta D. S., “Deep learning architecture LightOCT for diagnostic decision support using optical coherence tomography images of biological samples,” Biomed. Opt. Express 11(9), 5017–5031 (2020). 10.1364/BOE.395487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao X., Gan Y., Chang E., Hibshoosh H., Feldman S., Hendon C., “Visualization and tissue classification of human breast cancer images using ultrahigh-resolution OCT,” Lasers Surg. Med. 49(3), 258–269 (2017). 10.1002/lsm.22654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Xu Y., Boppart S. A., “Review of optical coherence tomography in oncology,” J. Biomed. Opt. 22(12), 1–23 (2017). 10.1117/1.JBO.22.12.121711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaitsev V., Matveev A., Matveev L., Sovetsky A., Hepburn M., Mowla A., Kennedy B., “Strain and elasticity imaging in compression optical coherence elastography: The two-decade perspective and recent advances,” J. Biophotonics 14(2), e202000257 (2021). 10.1002/jbio.202000257 [DOI] [PubMed] [Google Scholar]
- 28.Kennedy B. F., McLaughlin R. A., Kennedy K. M., Chin L., Wijesinghe P., Curatolo A., Tien A., Ronald M., Latham B., Saunders C. M., Sampson D. D., “Investigation of optical coherence microelastography as a method to visualize cancers in human breast tissue,” Cancer Res. 75(16), 3236–3245 (2015). 10.1158/0008-5472.CAN-14-3694 [DOI] [PubMed] [Google Scholar]
- 29.Allen W. M., Chin L., Wijesinghe P., Kirk R. W., Latham B., Sampson D. D., Saunders C. M., Kennedy B. F., “Wide-field optical coherence micro-elastography for intraoperative assessment of human breast cancer margins,” Biomed. Opt. Express 7(10), 4139–4153 (2016). 10.1364/BOE.7.004139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy K. M., Zilkens R., Allen W. M., Foo K. Y., Fang Q., Chin L., Sanderson R. W., Anstie J., Wijesinghe P., Curatolo A., Tan H. E. I., Morin N., Kunjuraman B., Yeomans C., Chin S. L., DeJong H., Giles K., Dessauvagie B. F., Latham B., Saunders C. M., Kennedy B. F., “Diagnostic accuracy of quantitative micro-elastography for margin assessment in breast-conserving surgery,” Cancer Res. 80(8), 1773–1783 (2020). 10.1158/0008-5472.CAN-19-1240 [DOI] [PubMed] [Google Scholar]
- 31.Fang Q., Frewer L., Zilkens R., Krajancich B., Curatolo A., Chin L., Foo K. Y., Lakhiani D. D., Sanderson R. W., Wijesinghe P., Anstie J. D., Dessauvagie B. F., Latham B., Saunders C. M., Kennedy B. F., “Handheld volumetric manual compression-based quantitative microelastography,” J. Biophotonics 13(6), e201960196 (2020). 10.1002/jbio.201960196 [DOI] [PubMed] [Google Scholar]
- 32.Gubarkova E. V., Sovetsky A. A., Zaitsev V. Y., Matveyev A. L., Vorontsov D. A., Sirotkina M. A., Matveev L. A., Plekhanov A. A., Pavlova N. P., Kuznetsov S. S., Vorontsov A. Y., Zagaynova E. V., Gladkova N. D., “OCT-elastography-based optical biopsy for breast cancer delineation and express assessment of morphological/molecular subtypes,” Biomed. Opt. Express 10(5), 2244–2263 (2019). 10.1364/BOE.10.002244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plekhanov A. A., Sirotkina M. A., Sovetsky A. A., Gubarkova E. V., Zaitsev V. Y., “Histological validation of in vivo assessment of cancer tissue inhomogeneity and automated morphological segmentation enabled by optical coherence elastography,” Sci. Rep. 10(1), 11781 (2020). 10.1038/s41598-020-68631-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirotkina M. A., Gubarkova E. V., Plekhanov A. A., Sovetsky A. A., Elagin V. V., Matveyev A. L., Matveev L. A., Kuznetsov S. S., Zagaynova E. V., Gladkova N. D., Zaitsev V. Y., “In vivo assessment of functional and morphological alterations in tumors under treatment using OCT-angiography combined with OCT-elastography,” Biomed. Opt. Express 11(3), 1365–1382 (2020). 10.1364/BOE.386419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gubarkova E. V., Kiseleva E. B., Sirotkina M. A., Vorontsov D. A., Gladkova N. D., “Diagnostic accuracy of cross-polarization OCT and OCT-elastography for differentiation of breast cancer subtypes: Comparative study,” Diagnostics 10(12), 994 (2020). 10.3390/diagnostics10120994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyazawa A., Makita S., Li E., Yamazaki K., Kobayashi M., Sakai S., Yasuno Y., “Polarization-sensitive optical coherence elastography,” Biomed. Opt. Express 10(10), 5162–5181 (2019). 10.1364/BOE.10.005162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.South F. A., Chaney E. J., Marjanovic M., Adie S. G., Boppart S. A., “Differentiation of ex vivo human breast tissue using polarization-sensitive optical coherence tomography,” Biomed. Opt. Express 5(10), 3417–3426 (2014). 10.1364/BOE.5.003417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Xu Y., Mesa K. J., South F. A., Chaney E. J., Spillman D. R., Barkalifa R., Marjanovic M., Carney P. S., Higham A. M., Liu Z. G., Boppart S. A., “Complementary use of polarization-sensitive and standard OCT metrics for enhanced intraoperative differentiation of breast cancer,” Biomed. Opt. Express 9(12), 6519–6528 (2018). 10.1364/BOE.9.006519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adie S. G., Hillman T. R., Sampson D. D., “Detection of multiple scattering in optical coherence tomography using the spatial distribution of stokes vectors,” Opt. Express 15(26), 18033–18049 (2007). 10.1364/OE.15.018033 [DOI] [PubMed] [Google Scholar]
- 40.Ju M. J., Hong Y. J., Makita S., Lim Y., Kurokawa K., Duan L., Miura M., Tang S., Yasuno Y., “Advanced multi-contrast jones matrix optical coherence tomography for doppler and polarization sensitive imaging,” Opt. Express 21(16), 19412–19436 (2013). 10.1364/OE.21.019412 [DOI] [PubMed] [Google Scholar]
- 41.Lurie K. L., Angst R., Ellerbee A. K., “Automated mosaicing of feature-poor optical coherence tomography volumes with an integrated white light imaging system,” IEEE Trans. Biomed. Eng. 61(7), 2141–2153 (2014). 10.1109/TBME.2014.2316535 [DOI] [PubMed] [Google Scholar]
- 42.Pande P., Shrestha S., Park J., Serafino M. J., Gimenez-Conti I., Brandon J. L., Cheng Y., Applegate B. E., Jo J. A., “Automated classification of optical coherence tomography images for the diagnosis of oral malignancy in the hamster cheek pouch,” J. Biomed. Opt. 19(8), 086022 (2014). 10.1117/1.JBO.19.8.086022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marvdashti T., Lian D., Aasi S. Z., Tang J. Y., Bowden A. K. E., “Classification of basal cell carcinoma in human skin using machine learning and quantitative features captured by polarization sensitive optical coherence tomography,” Biomed. Opt. Express 7(9), 3721–3735 (2016). 10.1364/BOE.7.003721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conners R. W., Harlow C. A., “A theoretical comparison of texture algorithms,” IEEE Trans. Pattern Anal. Machine Intell. PAMI-2(3), 204–222 (1980). 10.1109/TPAMI.1980.4767008 [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Allende P. B., Amygdalos I., Dhanapala H., Goldin R. D., Elson D. S., “Morphological analysis of optical coherence tomography images for automated classification of gastrointestinal tissues,” Biomed. Opt. Express 2(10), 2821–2836 (2011). 10.1364/BOE.2.002821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gnanadesikan R., Methods for Statistical Data Analysis of Multivariate Observations (Wiley, 1977). [Google Scholar]
- 47.Peng H., Long F., Ding C., “Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy,” IEEE Trans. Pattern Anal. Machine Intell. 27(8), 1226–1238 (2005). 10.1109/TPAMI.2005.159 [DOI] [PubMed] [Google Scholar]
- 48.Kittler J., “Feature selection and extraction,” in Handbook of Pattern Recognition and Image Processing,(Elsevier, 1986), pp.59–83. [Google Scholar]