Abstract
Coronavirus Disease 2019 (COVID-19), although most commonly demonstrates respiratory symptoms, but there is a growing set of evidence reporting its correlation with the digestive tract and faeces. Interestingly, recent studies have shown the association of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection with gastrointestinal symptoms in infected patients but any sign of respiratory issues. Moreover, some studies have also shown that the presence of live SARS-CoV-2 virus in the faeces of patients with COVID-19. Therefore, the pathophysiology of digestive symptoms associated with COVID-19 has raised a critical need for comprehensive investigative efforts. To address this issue we have developed a bioinformatics pipeline involving a system biological framework to identify the effects of SARS-CoV-2 messenger RNA expression on deciphering its association with digestive symptoms in COVID-19 positive patients. Using two RNA-seq datasets derived from COVID-19 positive patients with celiac (CEL), Crohn’s (CRO) and ulcerative colitis (ULC) as digestive disorders, we have found a significant overlap between the sets of differentially expressed genes from SARS-CoV-2 exposed tissue and digestive tract disordered tissues, reporting 7, 22 and 13 such overlapping genes, respectively. Moreover, gene set enrichment analysis, comprehensive analyses of protein–protein interaction network, gene regulatory network, protein–chemical agent interaction network revealed some critical association between SARS-CoV-2 infection and the presence of digestive disorders. The infectome, diseasome and comorbidity analyses also discover the influences of the identified signature genes in other risk factors of SARS-CoV-2 infection to human health. We hope the findings from this pathogenetic analysis may reveal important insights in deciphering the complex interplay between COVID-19 and digestive disorders and underpins its significance in therapeutic development strategy to combat against COVID-19 pandemic.
Keywords: SARS-CoV-2, COVID-19, digestive disorders, differentially expressed genes, functional enrichment, gene regulatory networks
Introduction
On the 11 March 2020, the World Health Organization (WHO) characterized the novel Coronavirus disease 2019 (COVID-19) resulting from the highly contagious Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) as a pandemic, which is, at present, still a major public health concern around the globe. According to the WHO report, there have been globally 78 299 811 confirmed cases of COVID-19 with more than 1.7 million deaths as of 22 December 2020 [1, 2]. Although at the early stage of the outbreak, the commonly reported symptoms in COVID-19 were related to the respiratory tract, recent data show that the digestive system of the human body might also be affected by SARS-CoV-2 infection. In one study, 204 COVID-19 patients were analyzed, and it was found that 103 patients presented symptoms related to the digestive system such as lack of appetite, diarrhoea, vomiting and abdominal pain. Although most patients reported fever or respiratory symptoms, for six patients, only digestive related side effects were presented amid the entire course of SARS-CoV-2 infection [3–5]. In another study, 1099 COVID-19 patients were analyzed from different hospitals of China, and it was observed that 55 of them (5.6%) had nausea or vomiting, or both, and 42 (3.8%) had diarrhoea [6, 7]. Jin and colleagues performed a similar study with COVID-19 patients and found that patients with SARS-CoV-2 with gastrointestinal (GI) symptoms such as diarrhoea, nausea and vomiting. They found 28% of those with GI symptoms did not have respiratory symptoms [8, 9]. In another investigation, the authors considered 95 SARS-CoV-2 infected patients and observed 58 cases (61.1%) of explicit GI symptoms with diarrhoea (24.2%), nausea (17.9%), vomiting (4.2%) [10]. In [11], the authors report that nearly 10% of SARS-CoV-2 infected patients presented with GI symptoms without respiratory symptoms. Several studies show that the presence of SARS-CoV-2 messenger RNA (mRNA) on stool samples, which indicates the relevance of GI tract symptoms with COVID-19 [12–15]. These findings indicate that the faecal–oral transmission could be an additional route for the spread of SARS-CoV-2 [16]. The characteristics of GI tract symptoms in COVID-19 are more deceptive than that of the respiratory tract symptoms. Some patients might have only GI symptoms amid the total course of infection, and some keep on shedding the infection in faeces regardless of respiratory samples testing negative.
However, the mechanism of how SARS-CoV-2 infection induces GI symptoms is still not elucidated and evidence for whether the infection can be transmitted through the GI tract other than the respiratory tract is yet to be examined. SARS-CoV-2 uses angiotensin-converting enzyme-2 (ACE-2) as a viral receptor to enter host cells [17–19]. Expression of the ACE-2 gene was reported in varieties of human tissues including intestine, testis, kidneys, heart and lungs [20, 21]. In addition, ACE-2 is a crucial regulator of intestinal inflammation [22], and an investigation on single-cell RNA sequencing data reveals that ACE-2 is highly expressed in the small intestine especially in proximal and distal enterocytes [23]. In another analysis on single-cell transcriptomes of lung, oesophagus, gastric, ileum and colon, Zhang et al. [24] found that, in addition to the presence of ACE-2, the SARS-CoV-2 virus also enters to the human body with the presence of the cellular serine protease, transmembrane protease serine-2 (TMPRSS-2). ACE-2 and TMPRSS-2 were not only coexpressed in lung alveolar type 2 cells and oesophageal upper epithelial and gland cells but also highly expressed in the ileum and colon. This indicates that SARS-CoV-2 can attack the enterocytes of the GI tract. In the oesophagus, ACE-2 was highly expressed in the upper and stratified epithelial cells. ACE-2 is more highly expressed in the ileum and colon than the lung [24].
The presence of SARS-CoV-2 on faeces, the interaction of ACE-2 and TMPRSS-2 and several GI symptoms suggest that further studies are required in order to understand the complex relevance of COVID-19 with the human digestive system. Since the GI tract is one of the crucial components of the digestive system, we have chosen three GI disorders in order to find the genetic relevance between COVID-19 and digestive disorder. In this work, we have considered celiac disease, Crohn’s disease and ulcerative colitis. Celiac disease (CEL) [25, 26] is an autoimmune condition that is also a digestive disorder that damages the small intestine’s lining and prevents it from absorbing some nutrients. The intestinal damage often causes GI problems such as diarrhoea, fatigue, weight loss, abdominal pain, nausea and vomiting, constipation, bloating and anaemia and can lead to serious complications. Crohn’s (CRO) and ulcerative colitis (ULC) belong to a group of conditions known as inflammatory bowel diseases (IBD) [27]. Crohn’s disease is a chronic inflammatory condition of the GI tract. Crohn’s disease causes inflammation in part of the digestive system and can affect any part of the GI tract from the mouth to the anus, but most commonly affects the end of the small intestine (ileum) and the beginning of the colon. Symptoms inside the GI tract often include abdominal pain, diarrhoea (with blood if inflammation is severe), fever, abdominal distension and weight loss [28]. Ulcerative colitis causes long-lasting inflammation and ulcers (sores) in the GI tract. Ulcerative colitis affects the innermost lining of the large intestine (colon) and rectum. The symptoms of active disease are abdominal pain, diarrhoea mixed with blood, weight loss, fever and anaemia [29, 30]. The exact causes of these diseases remain unknown, however, since the symptoms of these diseases are observed in COVID-19 samples, in this work, we have used the mRNA expression analysis in order to understand the complex interaction between COVID-19 and the digestive disorders. In other words, we have performed infectome and diseasome network analysis in order to discover the underlying genes and the ways these genes are expressed from the perspective of the interaction between COVID-19 and digestive disorders. We have used a statistical method to determine differentially expressed genes (DEGs) and used the shared DEGs to determine protein–protein interaction (PPI), gene regulatory networks (GRN) and protein–chemical interactions. We have also performed gene set enrichment analysis (GSEA) for discovering cell signalling pathways and ontology shared by common DEGs.
Materials and Methods
Overview of the workflow
Bioinformatics and system biology approaches are common to identify and analyze the comorbidity complexities [31–37]. Datasets required in this work have been identified and collected at the initial phase, which is detailed in the following subsection. Gene expression analysis has been performed in order to identify the DEGs from each of the datasets. Next, the common DEGs between COVID-19 and the other three disorders have been identified. These common DEGs are used to discover their PPIs and to perform GSEA to identify enriched cell signalling pathways and functional Gene Ontology (GO) terms. Next, the same set of common DEGs are used to discover two types of GRNs: DEGs–micro RNAs (miRNA) network and DEGs–transcription factors (TFs) network. Finally, protein–chemical compound interactions have also been discovered for the common DEGs.
Gene expression dataset
In this work, we have considered two RNA-Seq datasets of SARS-CoV-2 infection on the human body. Samples of the first dataset are taken from whole blood and peripheral blood mononuclear cells (PBMCs) from three SARS-CoV-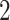 patients and three healthy individuals [38]. The second dataset (E-MTAB-8871) is taken from European Bioinformatics Institute Array Express Database [39], where the samples were collected from whole blood cells of COVID-
patients and three healthy individuals [38]. The second dataset (E-MTAB-8871) is taken from European Bioinformatics Institute Array Express Database [39], where the samples were collected from whole blood cells of COVID-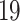 patients and healthy individuals by analyzing the immune responses using the NanoString Human Immunology Panel. We have used DESeq2 R package [40] to analyze the RNA-Seq data for SARS-CoV-2. For the digestive disorders, we have collected microarray data from the Gene Expression Omnibus (GEO), National Center for Biotechnology Information [41]. The dataset with GEO accession number of GSE113469 is considered in this work for celiac disease. The dataset investigates changes in the whole-genome profile of the PBMC of 17 patients with celiac disease with respect to 20 healthy controls. The dataset for the Crohn’s disorder (GSE3365) represents a comparison of PBMC transcriptional profiles between 42 healthy individuals and 59 patients with Crohn’s disease. The dataset with GEO accession number of GSE3365 is chosen for ulcerative colitis. In this dataset, transcriptional profiles in PBMCs from 42 healthy individuals and 26 ulcerative colitis patients were analyzed by hybridization to microarrays interrogating more than 22 000 sequences. The alterations in transcript levels observed by microarray were verified by real-time polymerase chain reaction.
patients and healthy individuals by analyzing the immune responses using the NanoString Human Immunology Panel. We have used DESeq2 R package [40] to analyze the RNA-Seq data for SARS-CoV-2. For the digestive disorders, we have collected microarray data from the Gene Expression Omnibus (GEO), National Center for Biotechnology Information [41]. The dataset with GEO accession number of GSE113469 is considered in this work for celiac disease. The dataset investigates changes in the whole-genome profile of the PBMC of 17 patients with celiac disease with respect to 20 healthy controls. The dataset for the Crohn’s disorder (GSE3365) represents a comparison of PBMC transcriptional profiles between 42 healthy individuals and 59 patients with Crohn’s disease. The dataset with GEO accession number of GSE3365 is chosen for ulcerative colitis. In this dataset, transcriptional profiles in PBMCs from 42 healthy individuals and 26 ulcerative colitis patients were analyzed by hybridization to microarrays interrogating more than 22 000 sequences. The alterations in transcript levels observed by microarray were verified by real-time polymerase chain reaction.
Identification of DEGs
Several statistical operations have been performed on the datasets in order to determine the DEGs. The limma (Linear Models for Microarray Analysis) R package has been used to perform statistical tests for identifying DEGs [37, 42]. In addition, the Benjamini–Hochberg false discovery rate method is used to provide a good balance between the discovery of statistically significant genes and the limitation of false positives. In this work, genes having adjusted P-value 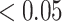 and absolute value of
and absolute value of 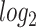 fold-change
fold-change 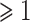 are considered as DEGs. We have also identified the DEGs from all the five datasets under consideration in this study: two for COVID-19 samples and three for digestive disorders. We have also identified the DEGs that are common in each of the COVID-19 datasets and digestive disorders. We have combined two sets of DEGs: (1) DEGs that are common in SARS-CoV-2 PBMC and digestive disorders and (2) DEGs that are common between SARS-CoV-2 immune and digestive disorders. In this article, we use the term combined DEGs’ to refer to the collection of these two sets of DEGs, which have been used in the downstream bioinformatics analyses.
are considered as DEGs. We have also identified the DEGs from all the five datasets under consideration in this study: two for COVID-19 samples and three for digestive disorders. We have also identified the DEGs that are common in each of the COVID-19 datasets and digestive disorders. We have combined two sets of DEGs: (1) DEGs that are common in SARS-CoV-2 PBMC and digestive disorders and (2) DEGs that are common between SARS-CoV-2 immune and digestive disorders. In this article, we use the term combined DEGs’ to refer to the collection of these two sets of DEGs, which have been used in the downstream bioinformatics analyses.
PPI analysis
We have constructed PPI networks using the combined DEGs that are common between COVID-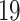 and digestive disorders. PPI analysis provides insight among the proteins and their association. We have identified protein subnetworks using STRING [43]—a protein interactome database. Gene clusters have been identified using the Markov cluster algorithm (MCL). We have used Cytoscape [44, 45] in order to generate the hub proteins network.
and digestive disorders. PPI analysis provides insight among the proteins and their association. We have identified protein subnetworks using STRING [43]—a protein interactome database. Gene clusters have been identified using the Markov cluster algorithm (MCL). We have used Cytoscape [44, 45] in order to generate the hub proteins network.
Functional enrichment analysis
To conduct the functional enrichment analysis with the combined DEGs, we have used Enrichr [46] with Fisher’s exact test. This overrepresentation analysis identified a set of enriched cell signalling pathways and functional GO terms that reveal the biological significance of the identified DEGs. We have merged the signalling pathways from six libraries in Enrichr including BioPlanet, BioCarta, KEGG, Panther, Reactome and WikiPathways. The duplicate pathways are removed, and only the significant pathways for which the adjusted P-value is less than 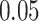 are considered. For functional GO terms, we have considered GO biological process (2018) dataset in Enrichr and determined the significant GO terms with adjusted P-value
are considered. For functional GO terms, we have considered GO biological process (2018) dataset in Enrichr and determined the significant GO terms with adjusted P-value 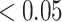 .
.
GRN analysis
GRN analysis includes the discovery of DEG–miRNA and TF–DEG interaction networks. The shared dysregulated genes are used to identify DEG–miRNA and TF–DEG networks using Network Analyst platform [47]. TarBase [48] and miRTarBase [49] databases are used for discovering DEG–miRNA interaction networks. JASPAR [50] database is used to identify TF–DEG interaction network. The networks are filtered with the betweenness value of 100.
Protein–Chemical compound analysis
Protein–chemical compound analysis identifies the chemical agents involved in the interaction of protein in comorbidity. In this analysis, we have identified protein–chemical interactions using the enriched gene (common DEGs) that COVID-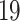 shares with digestive disorders. We have used Network Analyst [47] to identify protein–chemical interactions using the Comparative Toxicogenomics Database [51].
shares with digestive disorders. We have used Network Analyst [47] to identify protein–chemical interactions using the Comparative Toxicogenomics Database [51].
Result
Table 1.
A summary of experiments performed in this work to discover the comorbidity complexities between COVID-19 and digestive disorders
| Experiments | Input | Materials/methods | Presented in | Findings |
|---|---|---|---|---|
| Identification of DEG | Datasets of disorders with a list of genes | condition used: adjusted p-value 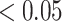 and absolute value of and absolute value of 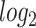 fold-change fold-change 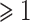
|
Volcano plots, Venn diagrams, Heatmaps | List of DEGs |
| PPI | Shared DEGs between COVID-19 and digestive disorders | STRING [43] | PPI interaction network | Interaction among proteins |
| Cell signalling pathways | Shared DEGs between COVID-19 and each of the digestive disorders | Enricher [46] | Bubble plots | List of significant signalling pathways |
| GO | Shared DEGs between COVID-19 and each of the digestive disorders | Enricher [46] | Bubble plots | List of significant GO pathways |
| DEGs–miRNA interaction | Shared DEGs between COVID-19 and each of the digestive disorders | Network Analyst [47] | Networks of DEGs–miRNA | Interactions between DEGs and miRNAs |
| DEGs–TFs interaction | Shared DEGs between COVID-19 and each of the digestive disorders | Network Analyst [47] | Networks of DEGs–TFs | Interactions between DEGs and TFs |
| Protein–chemical interaction | Shared DEGs between COVID-19 and each of the digestive disorders | Network Analyst [47] | Networks of proteins–chemicals | Interactions between proteins-chemicals agents |
Identifying genetic relevance between SARS-CoV-2 infection and digestive system
We have analyzed whole blood of SARS-CoV-2 infected patients and identified 1290 genes that are differentially expressed as compared with healthy control. Similarly, we have identified DEGs from the digestive disorders and found 108, 357 and 267 dysregulated genes for celiac, Crohn’s and ulcerative colitis disorders, respectively. The volcano plots presented in Figure 1 shows the DEGs for digestive disorders with the red dots. The number of shared DEGs between COVID-19 and three digestive disorders are presented in the Venn diagram shown in Figure 1D. COVID-19 shared 5, 11 and 13 dysregulated genes with celiac, Crohn’s and ulcerative colitis. The five shared DEGs between celiac and COVID-19 are KLRG1, NLRP3, IRAK3, MAFB and DDIT4. The family of Chemokine (C-X-C motif) genes, such as CXCL1, CXCL5, CLXL8, CXCR1, are highly expressed in the interaction between COVID-19 and Crohn’s disorder. We have found that 3 out of 11 shared genes between ulcerative colitis and COVID-19 are immunoglobulin-related DEGs such as IGKC, IGLJ3 and IGHA1. The DEGs shared between COVID-19 and digestive conditions and their relationships from the perspective of adjusted P-value and 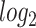 fold-change are presented in the heatmaps shown in Figure 1E and F, respectively.
fold-change are presented in the heatmaps shown in Figure 1E and F, respectively.
Figure 1.
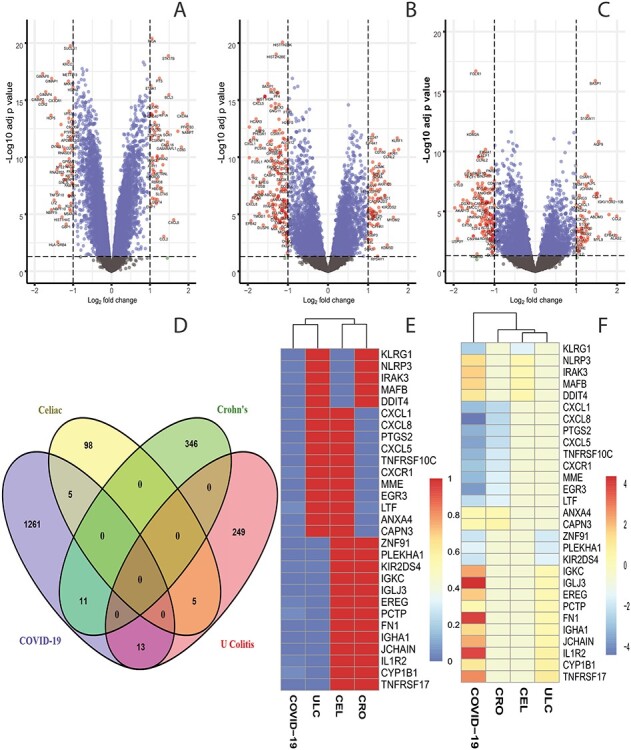
Red dots presented in volcano plots as shown in (A, B,C) represent the significant DEGs of celiac, Crohn’s and ulcerative colitis, respectively. The criteria chosen to be considered as DEGs are (i) absolute value of 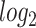 fold-change
fold-change 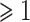 (ii) adjusted P-value <
(ii) adjusted P-value <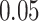 . The Venn diagram in (D) shows the number of DEGs and common DEGs among the conditions including COVID-19. Heatmaps show the relationships among common DEGs of different conditions based on (E) adjusted P-value and (F)
. The Venn diagram in (D) shows the number of DEGs and common DEGs among the conditions including COVID-19. Heatmaps show the relationships among common DEGs of different conditions based on (E) adjusted P-value and (F) 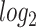 fold-change.
fold-change.
Identification of shared DEGs between the PBMC of immune response COVID-19 and digestive diseases
We have also analyzed the immune response from the PBMC of patients having SARS-CoV-2 infection and digestive comorbidity. Figure 2 shows the shared DEGs between the immune response of COVID-19 samples and digestive disorders. At first, we have identified 153 dysregulated genes in the COVID-19 immune response data. Next, we have discovered the overlapping genes between COVID-19 and other digestive conditions, which is depicted in the Venn diagram in Figure 2A. It is observed that the COVID-19 immune response sample does not share any DEGs with ulcerative colitis, whereas it shares 2 and 11 DEGs with celiac and Crohn’s disorders, respectively. The two overlapping DEGs between celiac and SARS-CoV-2 infection are CD79B and MBP. In the case of Crohn’s, two interleukin 1 receptor DEGs (IL1RL2 and IL1RAP), CD247, GNLY, EGR2 are some of the overlapping DEGs. We have also presented two heatmaps to show the associations among the overlapping DEGs. The heatmap presented in Figure 2B depicts the association between gene and diseases with adjusted P-value, and the heatmap shown in Figure 2C shows the relationship between gene and diseases considering 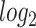 fold-change.
fold-change.
Figure 2.
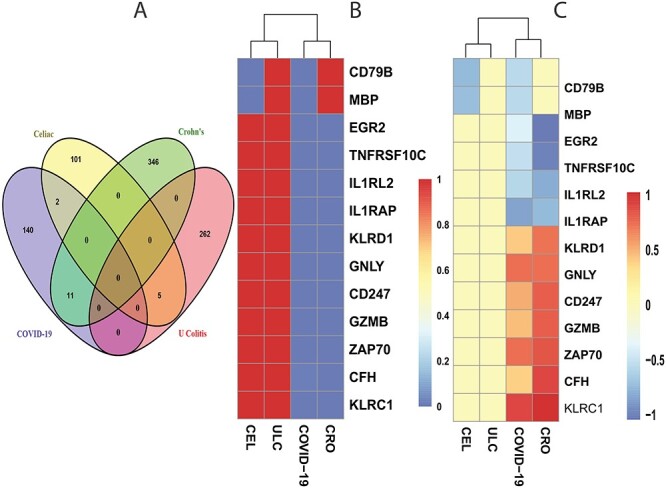
The Venn diagram in (A) shows the number of DEGs and overlapping DEGs among the disorders. The criteria chosen to be considered as DEGs are (i) 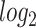 fold-change
fold-change 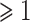 (ii) adjusted P-value <
(ii) adjusted P-value <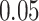 . Heatmaps show the relationships among overlapping DEGs of different disorders based on (B) adjusted P-value and (C)
. Heatmaps show the relationships among overlapping DEGs of different disorders based on (B) adjusted P-value and (C) 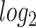 fold-change.
fold-change.
PPI analysis shows the hub proteins and the interaction among proteins
Next, We have employed PPI analysis with the shared DEGs between COVID-19 and digestive disorders. First, common DEGs from both whole-blood and immune response samples are used to construct the PPI network shown in Figure 3A. The rectangle boxes in the figure show the proteins involved in each of the digestive disorders and COVID-19. The edges show the interaction among the proteins, and thickness between the edges indicate the strength [43] of the relation among the proteins. The proteins with several connecting edges can be identified as hub proteins [52] [see Discussion for details]. Figure 3B shows a network of hub proteins. The network has been generated using Cytohubba [53] package of Cytoscape. As it is seen from this figure, CXCL8 has the highest connectivity among other proteins of the PPI network. The hub protein network displays the interaction of the hub proteins among each other. CXCL1, FN1, GZMB are also highly expressed among the hub proteins.
Figure 3.
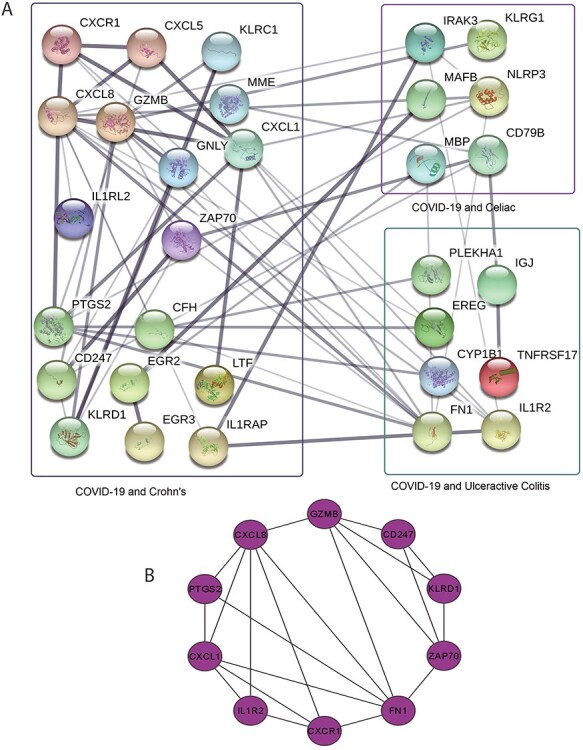
(A) PPI network of common DEGs in COVID-19 and digestive disorders. Square boxes represent the proteins shared by COVID-19 and corresponding disease. For example, the top right box represents the proteins that are common in the interaction between COVID-19 and celiac diseases. Proteins having several edges are highly expressed. (B) Hub protein network shows 10 hub proteins based on the number of interactions. CXCL8 has the highest interaction with other proteins.
Functional enrichment analysis identifies significant cell signalling pathways and GO terms
To identify significantly enriched cell signalling pathways and functional GO terms (biological process) with DEGs, we conducted functional enrichment test using Enrichr [46] tool. In this analysis, we have combined all the DEGs that have been discovered in the interaction of both COVID-19 PMBC and immune response cells with the digestive tract diseases and merged all the pathways from six pathway databases collected from Enrichr [46] libraries, i.e. BioPlanet, BioCarta, KEGG, Panther, Reactome and WikiPathways. We have considered the pathways for which adjusted P-value is less than 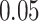 and plotted the top
and plotted the top 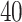 pathways for each of the digestive diseases in Figure 4. The pathways with higher logarithmic adjusted P-value are considered as significantly enriched. For example, the cytokine–cytokine receptor interaction pathway is the most significantly enriched pathway in the interaction between Crohn’s and COVID-19 as observed from Figure 4A. In addition, the family of interleukin pathways is most significantly enriched in Crohn’s–COVID comorbidity. We have also discovered some liver disorder-related pathways such as hepatitis C and hepatocellular carcinoma and hepatitis B. Immune system pathway also show significant enrichment, which represents the pathways between celiac and COVID-19 as show in Figure 4B. Several interleukin-1, Toll-like receptor (TLRs) pathways are also enriched in the celiac disorder. Moreover, the immune system, spinal cord injury, C-type lectin receptor signalling and interleukin-1 signalling pathways are common in both celiac and Crohn’s diseases with the influence of SARS-CoV-2 infection. Similar to the case of Crohn’s, the immune system pathway is also highly enriched in ulcerative colitis and COVID-19 pathways (Figure 4C). Scavenging of haeme from plasma, cytokine signalling in the immune system and FCERI-mediated MAPK activation are also among highly enriched pathways. In addition, we have observed several FCERI, ERBB2 and FGFR-related pathways in the association between COVID-19 and ulcerative colitis.
pathways for each of the digestive diseases in Figure 4. The pathways with higher logarithmic adjusted P-value are considered as significantly enriched. For example, the cytokine–cytokine receptor interaction pathway is the most significantly enriched pathway in the interaction between Crohn’s and COVID-19 as observed from Figure 4A. In addition, the family of interleukin pathways is most significantly enriched in Crohn’s–COVID comorbidity. We have also discovered some liver disorder-related pathways such as hepatitis C and hepatocellular carcinoma and hepatitis B. Immune system pathway also show significant enrichment, which represents the pathways between celiac and COVID-19 as show in Figure 4B. Several interleukin-1, Toll-like receptor (TLRs) pathways are also enriched in the celiac disorder. Moreover, the immune system, spinal cord injury, C-type lectin receptor signalling and interleukin-1 signalling pathways are common in both celiac and Crohn’s diseases with the influence of SARS-CoV-2 infection. Similar to the case of Crohn’s, the immune system pathway is also highly enriched in ulcerative colitis and COVID-19 pathways (Figure 4C). Scavenging of haeme from plasma, cytokine signalling in the immune system and FCERI-mediated MAPK activation are also among highly enriched pathways. In addition, we have observed several FCERI, ERBB2 and FGFR-related pathways in the association between COVID-19 and ulcerative colitis.
Figure 4.
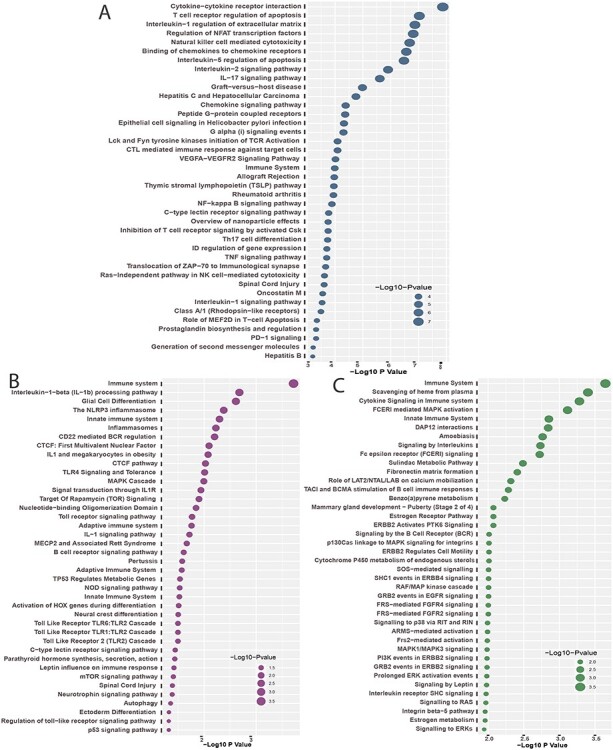
Top 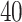 cell signalling pathways based on the adjusted p-values between COVID-
cell signalling pathways based on the adjusted p-values between COVID-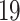 and digestive disorders. The pathways have been formed by combining the DEGs that are common in digestive disorder and both COVID-19 whole-blood and immune response samples. Figures (A, B and C) show the pathways for celiac, Crohn’s and ulcerative colitis, respectively.
and digestive disorders. The pathways have been formed by combining the DEGs that are common in digestive disorder and both COVID-19 whole-blood and immune response samples. Figures (A, B and C) show the pathways for celiac, Crohn’s and ulcerative colitis, respectively.
Furthermore, we have also conducted GO term enrichment using the same set of common DEGs. For this purpose, we have used the GO biological process (2018) database collected from Enrichr [46] libraries. The significantly enriched GO terms are identified if the enrichment yields high logarithmic value of adjusted P-value. Figure 5 displays the top 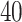 GO pathways of COVID-19 in relevance to digestive disorders. Positive regulation of neutrophil chemotaxis ontology term is highly enriched in Crohn’s disorder in presence of COVID-19 as observed from Figure 5A. Several neutrophil-related GO pathways are also highly enriched. Negative regulation of the immune system process and negative regulation of immune response are highly enriched GO pathways in celiac–COVID-19 interaction (Figure 5B). Also, the family of several interleukins related and T-helper 2 cell-related GO terms can be observed mostly between celiac and SARS-CoV-2 infection. In the case of ulcerative colitis, retina homeostasis is the most significantly enriched GO term. There are several cytokine-related GO terms are observed from Figure 5C.
GO pathways of COVID-19 in relevance to digestive disorders. Positive regulation of neutrophil chemotaxis ontology term is highly enriched in Crohn’s disorder in presence of COVID-19 as observed from Figure 5A. Several neutrophil-related GO pathways are also highly enriched. Negative regulation of the immune system process and negative regulation of immune response are highly enriched GO pathways in celiac–COVID-19 interaction (Figure 5B). Also, the family of several interleukins related and T-helper 2 cell-related GO terms can be observed mostly between celiac and SARS-CoV-2 infection. In the case of ulcerative colitis, retina homeostasis is the most significantly enriched GO term. There are several cytokine-related GO terms are observed from Figure 5C.
Figure 5.
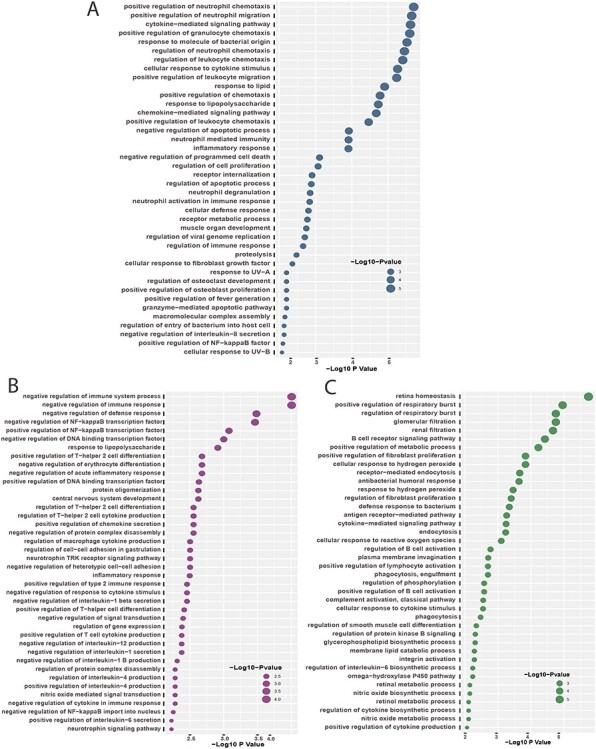
Top  GO terms based on the adjusted P-values between COVID-
GO terms based on the adjusted P-values between COVID- and digestive disorders. The GO terms have been identified with the combined DEGs that are common in digestive disorder and both COVID-19 whole-blood and immune response samples. (A–C) display the GO pathways for celiac, Crohn’s and ulcerative colitis, respectively.
and digestive disorders. The GO terms have been identified with the combined DEGs that are common in digestive disorder and both COVID-19 whole-blood and immune response samples. (A–C) display the GO pathways for celiac, Crohn’s and ulcerative colitis, respectively.
GRN analysis identifies networks of DEGs–miRNA and TF–gene interactions
We have used the common DEGs between COVID-19 and the other three digestive diseases. As we have done in the previous analysis, we have combined DEGs from both PBMC and immune response samples. The DEG–miRNA interactions network is displayed in Figure 6. The circles in the figure represent the dysregulated genes, and the squares represent the miRNAs. Lines connecting the nodes (circles or squares) of the networks represent the association among the nodes. Nodes in a network that connect several edges are termed significant nodes than others. For example, the network representing the DEG–miRNA association between COVID-19 and celiac is displayed in Figure 6A. In this figure, PTGS2, CXCL8, CXCL1, IL1RAP, MME, EGR3, CXCL5, EGR2 connect several miRNAs. In the case of COVID-19 and Crohn’s disorder interaction, we can observe DDIT4, MAFB, KLRG1, IRAK3 and MBP from Figure 6B, and among them DDIT4 and MAFB are more significant since these two DEGs have a higher degree (number of connecting edges) among the others. As far as miRNA is concerned, hsa-mir-124-3p is highly enriched. Other enriched miRNAs include hsa-mir-103a-3p, hsa-mir-16-5p, hsa-mir-107 and has-let-7b-5p. In ulcerative colitis–COVID-19 interaction (Figure 6C), the significant genes from the DEGs–miRNA network can be observed as PLBKHA1, CYP1B1, FN1, ZNF91 and PCTP. The dark brown squares represent significant miRNAs such as hsa-mir-7-5p, hsa-mir-30a-5p, hsa-mir-335-5p.
Figure 6.
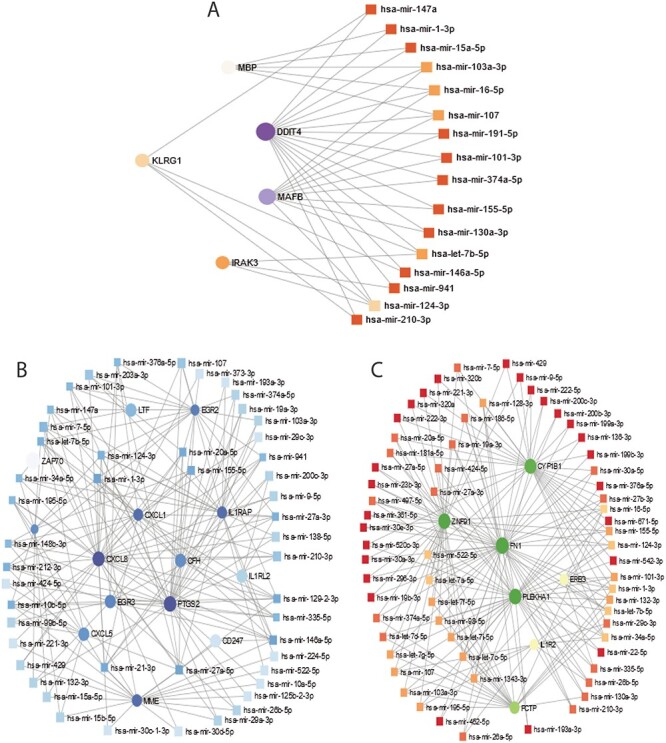
DEGs-miRNA networks of (A) Crohn’s and COVID-19 (B) celiac and COVID-19 and (C) ulcerative colitis and COVID-19. The networks are filtered with the betweenness value of 100. The circles represent DEGs and the squares represent miRNA. The highly expressed DEGs are presented with larger size and darker colour.
We have also identifiedTF–DEGs interaction networks of celiac, Crohn’s and ulcerative colitis with SARS-CoV-2 infection. The TF–DEGs networks are displayed in Figure 7. Circles and squares in the figure represent DEGs and TFs, respectively. Figure 7A displays TF–DEGs network between COVID-19 and celiac. As it can be seen, the significant DEGs in this network include EGR2, EGR3, CAPN3, ZAP70, CXCL5, KLRD1, LTF, CXCL1, GZMB. Significant TFs include FOXC1, GATA2, YY1, FOXL1, GATA3, STAT3. Figure 7B shows the TF–DEGs network between Crohn’s disorder and SARS-CoV-2 infection. The highly expressed gene in this network includes NLRP3, IRAK3, MBP, MAFB and KLRG1. FOXC1, GATA2, SREBF1 are among significant TFs as we observe from Figure 7B. In ulcerative colitis–COVID-19 interaction, the TF–DEGs network shows that FN1, PCTP, KIR2DS4, IGJ, PLEKHA1, IL1R2, CYP1B1 are among the highly expressed gene, and FOXC1, GATA2, FOXL1, YY1, NFKB1, PRDM1 are among significant TFs.
Figure 7.
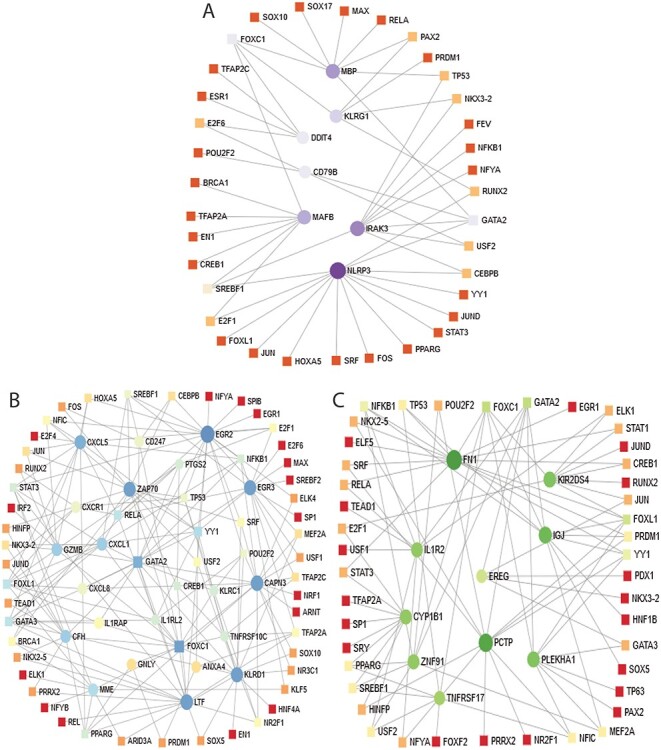
Association between COVID-19 and digestive tract disorders from the perspective of dysregulated gene and TFs. (A–C) displays the tf-gene networks of COVID-19 with Crohn’s, celiac and ulcerative colitis, respectively. The circles represent DEGs and the squares represent TFs. The highly expressed DEGs are presented with larger size and darker colour.
Protein–chemical compounds analysis identifies interactions between protein and chemical agents
Protein–chemical interaction is an important study to understand the protein functionality of proteins underpinning the molecular mechanisms within the cell, which may also help in the process of drug discovery. We have discovered the protein–chemical interaction networks of celiac, Crohn’s and ulcerative colitis with SARS-CoV-2 infection. The protein–chemical interaction networks are presented in Figure 7. Circles and squares in the figure represent proteins and chemical agents, respectively. Figure 8A displays the protein–chemical interaction network between celiac and SARS-CoV-2 infection on the human body. The significant proteins that can be identified from this network include DDTI4, MBP, MAFB and NLRP3. As far as chemical agents are concerned, Aflatoxin B1, Trichostatin A, Valproic acid are some of the highly enriched as observed. Figure 8B shows the protein–chemical network between Crohn’s disorder and SARS-CoV-2 infection. The highly expressed protein in this interaction includes PTGS2, CXCL8, CXCL1, CXCL5, CXCR1, EGR2 and CD247. In the case of ulcerative colitis and SARS-CoV-2 interaction, we have found CYP1B1, FN1, IL1R2 and EREG are highly expressed proteins as we can observe from Figure 8C.
Figure 8.
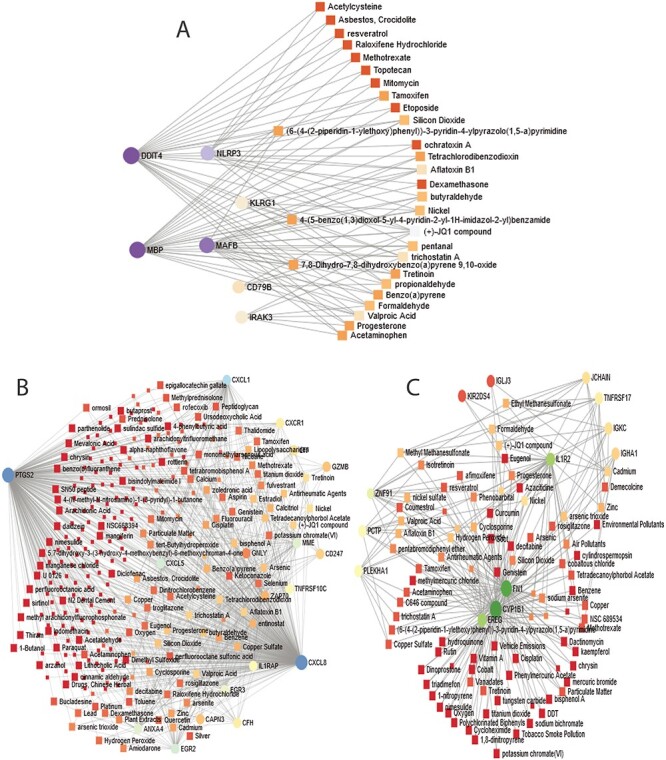
Association between COVID-19 and digestive tract disorders from the perspective of protein and chemical agents. (A–C) displays the protein–chemical networks of COVID-19 with celiac, Crohn’s and ulcerative colitis, respectively. The circles represent proteins and the squares represent chemical compounds. The highly expressed proteins are presented with larger size and darker colour. The squares having several edges are most expressed chemical agents.
Discussion
This work investigates the influences of SARS-CoV-2 infection on patients with digestive tract disorder. We have considered three digestive disorders and two datasets on SARS-CoV-2 infection and determined the DEGs from each of the datasets representing these diseases. Then, we have identified the shared DEGs between each of the digestive disorders and each of the datasets of SARS-CoV-2. In comparing the DEGs of celiac with two datasets (whole blood and immune response) of SARS-CoV-2 infection, we have identified 7 DEGs shared between celiac and COVID-19. The heatmaps in Figures 1 and 2 show the relationship between celiac and COVID-19 with respect to these DEGs. We have found IRAK3 (Interleukin 1 Receptor Associated Kinase 3), which belongs to the family of the interleukin-1 receptor-associated kinase protein is an important gene in mucosal immunity in the intestine, which acts as a cytoplasmic homeostatic mediator of inflammatory responses and inhibits signalling cascades downstream of myddosome complexes associated with TLRs. It has been reported that the alteration in IRAK3 is associated with susceptibility to asthma [54]. In addition, we have identified DDIT4 (DNA damage-inducible transcript 4) and MBP (myelin basic protein). DDIT4 plays an important role in DNA damage and energy stress, and the diseases associated with this gene include tuberous sclerosis, diabetes and bladder urothelial carcinoma [55], which has been demonstrated to be a proliferation-promoting and oncogenic protein in gastric cancer cells. Moreover, MBP is a significant gene that is linked with the nervous system [56]. Besides, the mutation in MBP is associated with the bone marrow and the immune system. One study shows suggest that MBP suppressed COX-2 expression play important role in the inhibition of growth and progression of gastric cancer indicating its important role in the immune defence system [57]. Altogether, these genes play vital roles in facilitating the immune system to fight when infected with SARS-CoV-2. We have identified 22 genes from both whole-blood and immune response samples of SARS-CoV-2 with Crohn’s disorder. We have discovered several shared genes from C-X-C Motif subfamilies of chemokine between Crohn’s and COVID-19 such as CXCL1, CXCL5, CXCL8 and CXCR1. Chemokines and their receptor families are important mediators for leukocyte migration to inflammatory sites such as those infected with SARS-CoV-2. The finding of these genes in our analysis also validates the idea that the family of C-X-C Motif genes might be the primary driver of the signature pathology observed in COVID-19 patients [58]. Alternate expression of CXCL1 is associated with the growth and progression of certain tumours. CXCL8 is interlinked with melanoma and respiratory distress. CXCR1 as well as CD247 and ZAP70 are responsible for several immunodeficiency disorders [59–61]. These indicate that the CXCL family proteins play a crucial role in inflammation during SARS-CoV-2 infection. EGR2 and EGR3 belong to the EGR family of C2H2-type zinc-finger proteins. Egr2 and Egr3 are vital for humoral immune tolerance in T cells. In SARS-CoV-2 patients, this might be functioning as a negative regulator of the T-cell activation so that there is a considerable balance between the activation and overactivation after infection. Recent studies show that the presence of GNLY can also be observed with COVID-19 patients having heart disease [31]. GNLY functions as a chemoattractant for T cells, monocytes and other inflammatory cells and stimulates several other cytokines and also helps in the recruitment of immune cells to the infection site as observed in SARS-CoV-2 infection. These genes are associated with bone disease, bipolar disorder and several neuronal developmental disorders [62, 63]. The presence of this gene might lead to further investigation towards the fact that SARS-CoV-2 affects the cardiovascular system of patients yielding greater fatality risks [64]. As far as ulcerative colitis is concerned, we have found several immunoglobulin-related DEGs such as IGKC, IGLJ3 and IGHA1, mutations in which might damage the human immune system [65]. We have also found KIR2DS4, which is a killer cell immunoglobulin-like receptor gene and responsible for other disorders besides GI diseases such as eye diseases, skin diseases and neuronal diseases. The finding of these genes presents evidence in connection with COVID-19 with skin and neuronal disorders [66–68]. Based on the number of shared signature genes, we have observed that SARS-CoV-2 infection affects mostly Crohn’s disease among the digestive disorders. Although Crohn’s diseases primarily affect the small intestine of a human body, this disease can attack any part of the digestive system unlike the other two conditions, which generally affect specific parts of the human digestive system.
With these dysregulated genes, we have conducted the PPI network analyses. Here, proteins with a minimum degree of 5, i.e. proteins that are connected to at least five edges are considered as hub proteins in the PPI network. As observed from the Figure 3, NLRP3 is the only hub protein for celiac. FN1 and IL1R2 can be considered hub proteins for ulcerative colitis, and in the case of Crohn’s, we may consider CXCR1, CXCL1, CXCL8, GZMB, PTGS2 and CD247 as hub proteins. These proteins, as have been discussed earlier, are involved in several other disorders. We have also identified significantly enriched GO terms and signalling pathways with the DEGs. These pathways and GO terms might determine the linkage among genes to identify potential therapeutic interventions. For example, the influence of SARS-CoV-2 infection on Crohn’s disorder reveals several Cytokine related pathways including chemokine and interleukin. Cytokines are significant in defending against infections and in other immune response [69]. The dysregulation in Chemokine and Interleukin might lead to inflammation and immune disorders. Interleukin-related pathways are also most expressed in celiac comorbidity with SARS-CoV-2 as we observe from our analysis. Interleukin-1 is an inflammatory cytokine, which has diverse physiological functionality and plays a significant role in health and disease. However, once the normal functionality of the interleukin-1 and its family such as interleukin-1- are disrupted, they may significantly contribute to the pathogenesis of not only inflammatory disease but also malignancies [70]. The identified pathways also support the findings presented in [71]. Overall, the GO terms indicate the activation of immune system pathways that help in the development, activation, regulation, migration and response in both innate and adaptive immunity.
are disrupted, they may significantly contribute to the pathogenesis of not only inflammatory disease but also malignancies [70]. The identified pathways also support the findings presented in [71]. Overall, the GO terms indicate the activation of immune system pathways that help in the development, activation, regulation, migration and response in both innate and adaptive immunity.
We have discovered DEG–miRNA, TF–DEG and protein–chemical agents interaction networks in order to understand the significance of the DEGs shared by SARS-CoV-2 infection with digestive disorders. In the case of celiac, the most expressed DEGs in these networks include DDIT4, MAFB, MBP, KLRG1, IRAK3 and NLRP3. We have identified hsa-miR-124-3p as the most expressed mi-RNA, which is associated with IRAK3, KLRG1, MAFB and DDIT4 genes. The hsa-miR-124-3p is involved in tumour progression, and the lower expression of hsa-miR-124-3p is likely to be correlated with the poor survival of patients with hepatocellular carcinoma [72]. We have found GATA2, FOXC1 and SREBF1 among more expressed TFs that are associated with DEGs shared between celiac and COVID-19. These factors are linked with other severe diseases such as neural, fatty liver disease, cancer[73, 74]. We have identified several interactions between proteins and several crucial chemical agents such as Aflatoxin B1, Trichostatin A and Valproic acid. Aflatoxin B1 is a common contaminant in a variety of foods including peanuts, corn and other gluten-based products. This chemical agent is a toxic substance that can damage the liver and can be the primary cause of Hepatocellular Carcinoma (HCC) [75]. Valproic acid is also a treat to the liver of the human digestive system. This acid may cause serious birth defects and bring harm to the spinal cord and brain [76]. These results provide evidence on the pathways and ontology, which has been found in this work, as we have found pathways that are related to the liver, spinal cord injury and neural diseases. These three chemical components are also associated with the protein-chemical interaction of SARS-CoV-2 infection with IBD (Crohn’s and ulcerative colitis disorders). We have discovered several significant miRNAs in the interaction between Crohn’s and COVID-19 such as hsa-mir-1-3p, hsa-mir-124-3p, hsa-mir-129-2-3p. Studies have shown that miR-1-3p is known to be involved in heart diseases [31] and also plays significant roles in the development of human cancers such as lung cancer, bladder cancer or prostate cancer [77]. These two mi-RNAs are also available in the network of ulcerative colitis and COVID-19. Similar to the celiac disease, we have found GATA2, FOXC1, SREBF1 TFs in Crohn’s and ulcerative colitis. In addition, GATA3, PPARG, NFKB1, FOXL1 are also some highly expressed TFs in IBD. FOXC1 and FOXL1 belong to the forkhead box (FOX) family of TFs. These two genes significantly regulate intestine functionality and also play important role in the pathogenesis of heart diseases [78]. Karban et al. [79] showed that increased expression of NFKB1 (nuclear factor kappa B subunit 1) is one of the important causes of ulcerative colitis, which is concordant with our findings. Besides, NFKB1 is associated with immune deficiency and renal diseases.
Conclusion
In this work, we have considered RNA-seq data of SARS-CoV-2 infection in human, and microarray datasets of three digestive disorders. This study focuses on the identification of the biomarkers between the association of SARS-CoV-2 and digestive disorders from the perspective of molecular and cellular levels. The result of this study has discovered several significant genes such as IRAK3, DDIT4, MBP, CXCR1, CXCL1, CXCL8, CD247, GNLY, EGR3, IGLJ3 and FN1 to name a few. Our result also reveals that the number of shared genes between Crohn’s and COVID-19 is greater than other two digestive disorders. We have provided gene expression analysis with the identified biomarker to discover the cell signalling pathways and GO terms. In addition, we have performed PPIs to identify the significant proteins and their relationships in COVID-19 patients’ bodies with the presence of disorder on the GI tract. We have also identified the hub proteins, and our result reveals CXCL8 is the most expressed protein based on the number of interactions with other proteins. Moreover, the GRN analysis identifies the involvement of potential TFs and miRNA, which highlights the risk factors associated with SARS-CoV-2 infection. The analysis of the interaction between proteins and chemical compounds reveals the presence of significant chemical agents. Our overall analysis discovers the involvement of biomarkers in other risk factors such as disorders of the cardiovascular, renal, brain and autoimmune, which provides the evidence on the influence of SARS-CoV-2 infection on different parts of the human body. We hope that the results found in this study facilitate significant insights that might help in developing novel/repurpose existing therapeutic strategies to combat COVID-19.
Key Points
This work focuses on the influences of SARS-CoV-2 infection on individuals having digestive disorders.
Using RNA-seq datasets derived from COVID-19 positive patients with celiac, Crohn’s and ulcerative colitis as digestive disorders, we have found a significant association with the digestive tract disordered tissues.
We employed global transcriptomic data to explore their shared expression profile and their co-expression, important biomarkers, regulatory networks (transcription factors and miRNAs) and signalling pathways that could influence each other’s clinical outcome.
The important genes, proteins and signalling pathways could be useful as therapeutic targets and molecular checkpoints to control the COVID-19 pandemic.
this study could aid in our understanding of COVID-19 progression due to prevalent health conditions and its further health impact.
Md Asif Nashiry received his PhD degree from the University of Lethbridge, Alberta, Canada in 2018. Currently, he is working as an associate professor at the Department of Computer Science and Engineering of the Jashore University of Science and Technology. His research areas of interest include machine learning, data science and bioinformatics.
Shauli Sarmin Sumi is working as an assistant professor at the Department of Computer Science and Engineering of Jashore University of Science and Technology. She received her MSc in Computer Science from the University of Lethbridge, Canada. She works in the areas of natural language processing, data mining and bioinformatics.
Mohammad Umer Sharif Shohan is a lecturer at the Department of Biochemistry and Molecular Biology of the University of Dhaka. He has worked in different organizations around the world including European Bioinformatics Institute (UK), International Research Center for Medical Sciences (Japan) and University of Queensland (Australia). His area of expertise is single-cell transcriptomics, mathematical modeling, machine learning and biostatistics.
Salem A. Alyami received the PhD degree from Monash University, Australia in biostatistics in 2017. He is an assistant professor in the School of Mathematics and Statistics, IMAMU Riyadh, since 2017, contributing/leading several grants in bioistatistics projects. Recently, he has appointed as the Dean of the Deanship of Scientific Research at IMAMU, Riyadh, KSA. His research interest includes, Bayesian networks, neural networks, Bayesian statistics, MCMC methods, applications of statistics in biology and medicine.
A.K.M. Azad received a PhD degree from Monash University, Australia in computational systems biology and biostatistics in 2017, followed by his first postdoctoral fellowship in the Faculty of IT, Monash University. He was a postdoctoral research associate in the AI Lab in the School of BABS, UNSW Sydney, contributing/leading several bioinformatics and computational biology projects. Recently, he has joined the University of Technology, Sydney as a Research Fellow for developing cutting-edge methodologies and tools for online inference of phylogenetic trees from COVID-19 sequences. His research interest includes AI/ML/DL, Bayesian statistics, MCMC, Bayesian network, bioinformatics and computational biology.
Mohammad Ali Moni is a research fellow and conjoint lecturer at the University of New South Wales, Australia. He received his PhD in clinical bioinformatics and machine learning from the University of Cambridge. His research interests encompass artificial intelligence, machine learning, data science, medical image processing and clinical bioinformatics.
Contributor Information
Md Asif Nashiry, Department of Computer Science and Engineering, Jashore University of Science and Technology, Jashore, Bangladesh.
Shauli Sarmin Sumi, Department of Computer Science and Engineering, Jashore University of Science and Technology, Jashore, Bangladesh.
Mohammad Umer Sharif Shohan, Department of Biochemistry and Molecular Biology,University of Dhaka, Dhaka-1000, Bangladesh.
Salem A Alyami, Department of Mathematics and Statistics, Faculty of Science, Imam Mohammad Ibn Saud Islamic University (IMSIU), Riyadh 13318, Saudi Arabia.
A K M Azad, iThree Institute, Faculty of Science, University Technology of Sydney, Australia.
Mohammad Ali Moni, WHO Collaborating Centre on eHealth, UNSW Digital Health, School of Public Health and Community Medicine, Faculty of Medicine, UNSW Sydney, Australia; Healthy Ageing Theme, The Garvan Institute of Medical Research, Darlinghurst, NSW 2010, Australia.
Author contributions statement
M.A.N. conceived and designed the study, M.A.N. conducted the experiments, performed the computational analysis, prepared the illustrations, and wrote the manuscript, S.S.S. contributed in computational analysis and manuscript writing, M.U.S.S, S.A.A and A.K.M.A contributed in critical revision, M.A.M. supervised the whole study. All authors approved the final version for submission.
Competing interests
The authors declare no conflict of interest.
References
- 1. World Health Organization . https://covid19.who.int/ (2020). (accessed 2020 Aug 21).
- 2. Uddin S, Imam T, Moni MA, et al. Onslaught of Covid-19: how did governments react and at what point of the crisis? Popul Health Manag 2020;24(1):13–19. [DOI] [PubMed] [Google Scholar]
- 3. Pan L, Mu M, Yang P, et al. Clinical characteristics of Covid-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol 2020;115(5):766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahamad MM, Aktar S, Rashed-al-Mahfuz M, et al. A machine learning model to identify early stage symptoms of SARS-COV-2 infected patients. Expert Syst Appl 2020;160:113661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aktar S, Talukder A, Ahamad MM, et al. Machine learning and meta-analysis approach to identify patient comorbidities and symptoms that increased risk of mortality in Covid-19 arXiv preprint arXiv:2008.12683. 2020. [DOI] [PMC free article] [PubMed]
- 6. Guan W-j, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uddin S, Imam T, Khushi M, et al. How do socio-demographic status and personal attributes influence adherence to Covid-19 preventive behaviours? medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (Covid-19) with gastrointestinal symptoms. Gut 2020;69:1002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taz TA, Ahmed K, Paul BK, et al. Network-based identification genetic effect of SARS-COV-2 infections to idiopathic pulmonary fibrosis (IPF) patients. Brief Bioinform 2020;22(2):1254–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin L, Jiang X, Zhang Z, et al. SARS-COV-2 infection. Gut 2020;69:997–1001. [DOI] [PubMed] [Google Scholar]
- 11. Mao R, Qiu Y, He JS, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with Covid-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2020;5(7):667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020;382(10):929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-COV-2. Gastroenterology 2020;158:1831–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W, Xu Y, Gao R, et al. Detection of SARS-COV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chowdhury UF, Sharif Shohan MU, Hoque KI, et al. A computational approach to design potential SIRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-COV-2. Genomics 2021;113:331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uddin S, Imam T, Ali Moni M. The implementation of public health and economic measures during the first wave of Covid-19 by different countries with respect to time, infection rate and death rate. In: 2021 Australasian Computer Science Week Multiconference, Dunedin, New Zealand, ACM, 2021, 1–8.
- 17. Hoffmann M, Kleine-Weber H, Schroeder S, et al. Sars-cov-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-COV-2 by full-length human ACE2. Science 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oany AR, Mia M, Pervin T, et al. Design of novel viral attachment inhibitors of the spike glycoprotein (S) of severe acute respiratory syndrome coronavirus-2 (SARS-COV-2) through virtual screening and dynamics. Int J Antimicrob Agents 2020;56(6):106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M-Y, Li L, Zhang Y, et al. Expression of the SARS-COV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 2020;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohammad Umer Sharif Shohan, A. S. M. Rubayet Ul Alam, Nadira Naznin Rakhi, et al. Onset, Transmission, Impact, and Management of COVID-19 Epidemic at Early Stage in SAARC Countries. Authorea. 2020. doi: 10.22541/au.159775079.90952648. [DOI] [Google Scholar]
- 22. Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012;487:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang W, Feng Z, Rao S, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut 2020;69:1141–3. [DOI] [PubMed] [Google Scholar]
- 24. Zhang H, Kang Z, Gong H, et al. Digestive system is a potential route of Covid-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020;69:1010–8. [Google Scholar]
- 25. Celiac Disease Foundation . https://celiac.org/about-celiac-disease/what-is-celiac-disease/ 2020. (13 April 2021, date last accessed).
- 26. Kochhar GS, Singh T, Gill A, et al. Celiac disease: managing a multisystem disorder. Cleve Clin J Med 2016;83:217–27. [DOI] [PubMed] [Google Scholar]
- 27. Duchmann R, Kaiser I, Hermann E, et al. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clinical & Experimental Immunology 1995;102:448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 2012;380:1590–605. [DOI] [PubMed] [Google Scholar]
- 29. Colitis-Pathophysiology U. Inflammatory bowel disease part i: ulcerative colitis–pathophysiology and conventional and alternative treatment options. Altern Med Rev 2003;8:247–83. [PubMed] [Google Scholar]
- 30. Uddin S, Imam T, Khushi M, et al. How did socio-demographic status and personal attributes influence compliance to Covid-19 preventive behaviours during the early outbreak in japan? Lessons for pandemic management. Personal Individ Differ 2021;110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nashiry A, Sarmin Sumi S, Islam S, et al. Bioinformatics and system biology approach to identify the influences of Covid-19 on cardiovascular and hypertensive comorbidities. Brief Bioinform 2021;22(2):1387–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moni MA, Liò P. comor: a software for disease comorbidity risk assessment. J Clin Bioinform 2014;4:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nain Z, et al. Pathogenetic profiling of Covid-19 and SARS-like viruses. Brief Bioinform 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moni MA, Liò P. How to build personalized multi-omics comorbidity profiles. Front Cell Dev Biol 2015;3:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Satu MS, Khan MI, Rahman MR, et al. Diseasome and comorbidities complexities of SARS-COV-2 infection with common malignant diseases. Brief Bioinform 2021;22(2):1415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moni MA, Quinn JM, Sinmaz N, et al. Gene expression profiling of SARS-COV-2 infections reveal distinct primary lung cell and systemic immune infection responses that identify pathways relevant in Covid-19 disease. Brief Bioinform 2020;22(2):1324–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moni MA, Rana HK, Islam MB, et al. A computational approach to identify blood cell-expressed Parkinson’s disease biomarkers that are coordinately expressed in brain tissue. Comput Biol Med 2019;113:103385. [DOI] [PubMed] [Google Scholar]
- 38. Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in Covid-19 patients. Emerging microbes & infections 2020;9:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goujon M, McWilliam H, Li W, et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res 2010;38:W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for Rna-seq data with deseq2. Genome Biol 2014;15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 2012;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2018;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 2011;27:431–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moni MA, Liò P. Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies. BMC Bioinform 2014;15:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xia EEG, Jianguo Hancock RE. Network analyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015;10:823–44. [DOI] [PubMed] [Google Scholar]
- 48. Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res 2011;40:D222–9. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang H-Y, Lin YCD, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 2019;48:D148–54. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fornes O, Castro-Mondragon JA, Khan A, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2019;48:D87–92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis AP, Grondin CJ, Johnson RJ, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res 2018;47(D1):D948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ekman D, Light S, Björklund ÅK, et al. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol 2006;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chin C-H, Chen S-H, Wu HH, et al. cytohubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pino-Yanes M, Sánchez-Machín I, Cumplido J, et al. IL-1 receptor–associated kinase 3 gene (irak3) variants associate with asthma in a replication study in the Spanish population. J Allergy Clin Immunol 2012;129:573–5. [DOI] [PubMed] [Google Scholar]
- 55. Pinto JA, Rolfo C, Raez LE, et al. In silico evaluation of DNA damage inducible transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci Rep 2017;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Campagnoni AT. Molecular biology of myelin proteins from the central nervous system. J Neurochem 1988;51:1–14. [DOI] [PubMed] [Google Scholar]
- 57. Hsu K-W, Hsieh RH, Wu CW, et al. Mbp-1 suppresses growth and metastasis of gastric cancer cells through COX-2. Mol Biol Cell 2009;20:5127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-COV-2 drives development of Covid-19. Cell 2020;181(5):1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Teruel M, McKinney C, Balsa A, et al. Association of cd247 polymorphisms with rheumatoid arthritis: a replication study and a meta-analysis. PLoS one 2013;8:e68295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walkovich K, Vander Lugt M. Zap70-related combined immunodeficiency. In: GeneReviews [Internet]. Seattle, WA: University of Washington, 2017. [Google Scholar]
- 61. Proost P, Struyf S, Loos T, et al. Coexpression and interaction of cxcl10 and cd26 in mesenchymal cells by synergising inflammatory cytokines: Cxcl8 and cxcl10 are discriminative markers for autoimmune arthropathies. Arthritis Res Ther 2006;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Safran M, Dalah I, Alexander J, et al. GeneCards version 3: the human gene integrator. Database 2010;2010:baq020. 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tewary P, Yang D, Rosa G, et al. Granulysin activates antigen-presenting cells through TLR4 and acts as an immune alarmin. Blood 2010;116:3465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng Y-Y, Ma Y-T, Zhang J-Y, et al. Covid-19 and the cardiovascular system. Nat Rev Cardiol 2020;17:259–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dunn-Walters DK, Isaacson PG, Spencer J. Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) b cells suggests that the MGZ of human spleen is a reservoir of memory b cells. J Exp Med 1995;182:559–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y-C, Bai W-Z, Hashikawa T. The neuroinvasive potential of SARS-COV2 may play a role in the respiratory failure of Covid-19 patients. J Med Virol 2020;92:552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu Y, Xu X, Chen Z, et al. Nervous system involvement after infection with Covid-19 and other coronaviruses. Brain Behav Immun 2020;87:18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bouaziz J, Duong TA, Jachiet M, et al. Vascular skin symptoms in Covid-19: a French observational study. J Eur Acad Dermatol Venereol 2020;34(9):e451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luster AD. Chemokines-chemotactic cytokines that mediate inflammation. N Engl J Med 1998;338:436–45. [DOI] [PubMed] [Google Scholar]
- 70. Kaneko N, Kurata M, Yamamoto T, et al. The role of interleukin-1 in general pathology. Inflammation Regener 2019;39:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villapol S. Gastrointestinal symptoms associated with Covid-19: impact on the gut microbiome. Transl Res 2020;226:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Long H-D, Ma YS, Yang HQ, et al. Reduced hsa-mir-124-3p levels are associated with the poor survival of patients with hepatocellular carcinoma. Mol Biol Rep 2018;45:2615–23. [DOI] [PubMed] [Google Scholar]
- 73. Hahn CN, Chong CE, Carmichael CL, et al. Heritable gata2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet 2011;43:1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ray PS, Wang J, Qu Y, et al. Foxc1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res 2010;70:3870–6. [DOI] [PubMed] [Google Scholar]
- 75. McKean C, Tang L, Tang M, et al. Comparative acute and combinative toxicity of aflatoxin b1 and fumonisin b1 in animals and human cells. Food Sand Chem Toxicol 2006;44:868–76. [DOI] [PubMed] [Google Scholar]
- 76. Lammer EJ, Sever LE, Oakley GP, Jr. Valproic acid. Teratology 1987;35:465–73. [DOI] [PubMed] [Google Scholar]
- 77. Shang A, Yang M, Shen F, et al. Mir-1-3p suppresses the proliferation, invasion and migration of bladder cancer cells by up-regulating SFRP1 expression. Cell Physiol Biochem 2017;41:1179–88. [DOI] [PubMed] [Google Scholar]
- 78. Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci 2016;144:194–201. [DOI] [PubMed] [Google Scholar]
- 79. Karban AS, Okazaki T, Panhuysen CIM, et al. Functional annotation of a novel nfkb1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet 2004;13:35–45. [DOI] [PubMed] [Google Scholar]


