Fig. 1. Schematic of the modular molecular design pipeline.
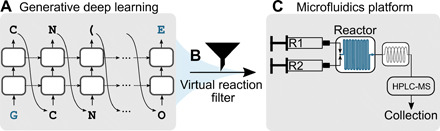
(A) A generative deep learning model (27) based on a long-short term memory network (34) was used to generate putative liver X receptor α agonists. The two-step network training procedure first trained the model on 656,070 compounds predicted as compatible with the microfluidics system and then fine-tuned this pretrained model with 40 known LXRα agonists. (B) The de novo generated molecules were filtered on the basis of their predicted synthesis route, using a set of 17 reactions specified in the SMARTS notation (“virtual reaction filter”). Molecular building blocks for synthesis were automatically retrieved from a commercial supplier catalog. (C) A total of 41 de novo designs were retained for synthesis on a microfluidics platform, which contained two syringes for the handling of reagents R1 and R2, a Cetoni Qmix element equipped with a Dean Flow microfluidic reactor chip, and a Rheodyne MRA splitter for automated sample transfer to an HPLC-MS system.
