Fig. 2. Automating de novo design with deep learning.
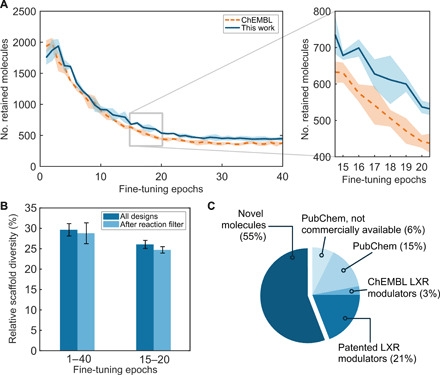
(A) Number of de novo designs retained by the virtual reaction filter depending on the pretraining (mean and SD over three replicates and 3000 sampled SMILES strings each). Compared to previous studies using bioactive molecules from ChEMBL (dashed lines) (9, 27, 38), this pretraining strategy (solid line) led to a larger number of compounds retained by the virtual reaction tool (P < 0.001, Kruskal-Wallis test), with up to 255 ± 97 more designs retained in each fine-tuning epoch. The epochs chosen for sampling are highlighted (epochs 15 to 20, gray rectangle). (B) Relative scaffold diversity (i.e., unique scaffolds/total number of scaffolds) of the de novo designs before and after applying the virtual reaction filter. No statistically significant difference in scaffold diversity was observed (Wilcoxon test, α = 0.05). (C) Analysis of 67 de novo designs retained for potential synthesis: 14 compounds were patented LXR modulators annotated in SureChEMBL or Reaxys (22%); 15 compounds existed in PubChem, of which 10 compounds are annotated as commercially available (15% of the total); 4 (6%) compounds lack vendor information; and 2 compounds (3%) were known LXR modulators annotated in ChEMBL27 [median inhibitory concentration (IC50)/EC50 ≤ 2 μM]. Thirty-seven compounds (55%) were not found in either PubChem, ChEMBL27, SciFinder, SureChEMBL, or Reaxys databases.
