Fig. 3. Ternary structure of Polθ on a DNA/RNA primer-template.
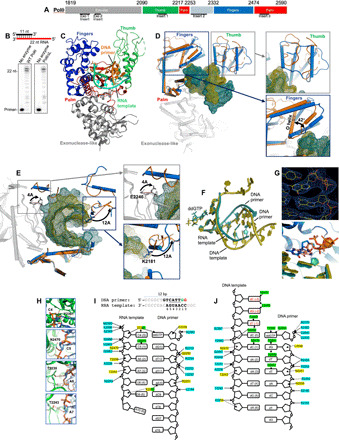
(A) Polθ polymerase. (B) DNA/RNA extension by Polθ and PolθΔL. (C) Structure of Polθ:DNA/RNA:ddGTP. (D) Superposition of Polθ:DNA/RNA (marine) and Polθ:DNA/DNA (orange, 4x0q). The fingers and thumb subdomains undergo reconfiguration. (E) Superposition of Polθ:DNA/RNA (marine) and Polθ:DNA/DNA (orange, 4x0q) highlighting a 12-Å shift of K2181 (blue box; thumb) and a 4.4-Å shift of E2246 (gray box; palm). (F) Superposition of nucleic acids and ddGTP from Polθ:DNA/RNA:ddGTP and Polθ:DNA/DNA:ddGTP structures. (G) Top: Electron density of ddGTP and 3′ primer terminus in Polθ:DNA/RNA structure. Bottom: Zoomed-in image of the superposition of active sites, illustrating a different conformation of ddGTP in the Polθ:DNA/RNA (blue) and Polθ:DNA/DNA (salmon) complexes. (H) Interactions between ribose 2′-hydroxyl groups of the RNA template and residues in the Polθ:DNA/RNA structure. Red dashed lines, hydrogen bonds. (I) DNA/RNA used for cocrystallization with Polθ and ddGTP (top). Strong electron density is present for four base pairs [nucleotides located at positions 2 to 5 (underlined) of the DNA/RNA] and two base pairs resulting from an incorporated ddGMP (2’,3’ dideoxyguanosine monophosphate) (green; position 1) and a bound unincorporated ddGTP (red; position 0) in the active site (top). Interactions between Polθ and nucleic acids in Polθ:DNA/RNA:ddGTP (bottom). Interactions between residues and phosphate backbone, sugar oxygen, or nucleobase are shown in blue, yellow, and green, respectively. Hydrogen bonds between Polθ and ribose 2′-hydroxyl groups are indicated (boxed residues). (J) Interactions between Polθ and nucleic acids in Polθ:DNA/DNA:ddGTP (4x0q). Color scheme identical to (I).
