Fig. 3. Impact of molecular-scale spatial control of two extracellular ligands on NK cell immune response.
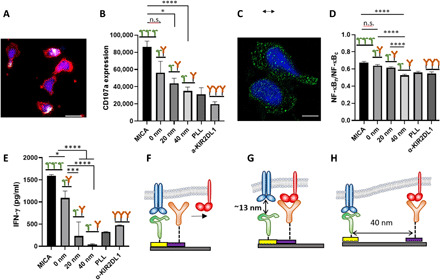
(A) NK cells stimulated on nanopattern of MICA continuous MICA film, respectively. The cells were stained with phalloidin for cytoskeleton (red), 4′,6-diamidino-2-phenylindole for nucleus (blue), and anti-CD107a (white). Scale bar, 10 μm. (B) Average amount of membrane-bound CD107a per cell on ligand patterns (n ≥ 60) compared to that of cells stimulated on control surfaces (n ≥ 60). Surfaces with continuous MICA provided the optimal condition for the activation of NK cells, whereas surfaces with poly-l-lysine and KIR2DL1 did not stimulate significant CD107a expression owing to the absence of activating ligands. Patterned arrays of MICA and anti-KIR2DL1 produced activation that depended on the gap between the ligands. (C) Nuclear factor κB (NF-κB) p65 staining (green) of activated NK cells. Scale bar, 5 μm. (D) Degree of NF-κB nuclear translocation on ligand patterns (n ≥ 60) compared to that of cells on control surfaces (n ≥ 30). (E) Interferon-γ (IFN-γ) release intensity. (F to H) Schemes of the possible mechanisms explaining the observed effect of the gap between the ligands in NK cell activation: (F) long ligand binds first, precluding binding to the colocalized shorter ligand; (G) short ligand binds first, precluding binding to the colocalized longer ligands; and (H) both ligands can bind their receptors owing to the spatial segregation. Data are representative of three experiments [means and SEM in (B), (D), and (E)]. In brackets, n represents the number of imaged regions. The results were considered to be significantly different for P < 0.05. *P < 0.05, ***P < 0.001, ****P < 0.0001. n.s., not significant.
