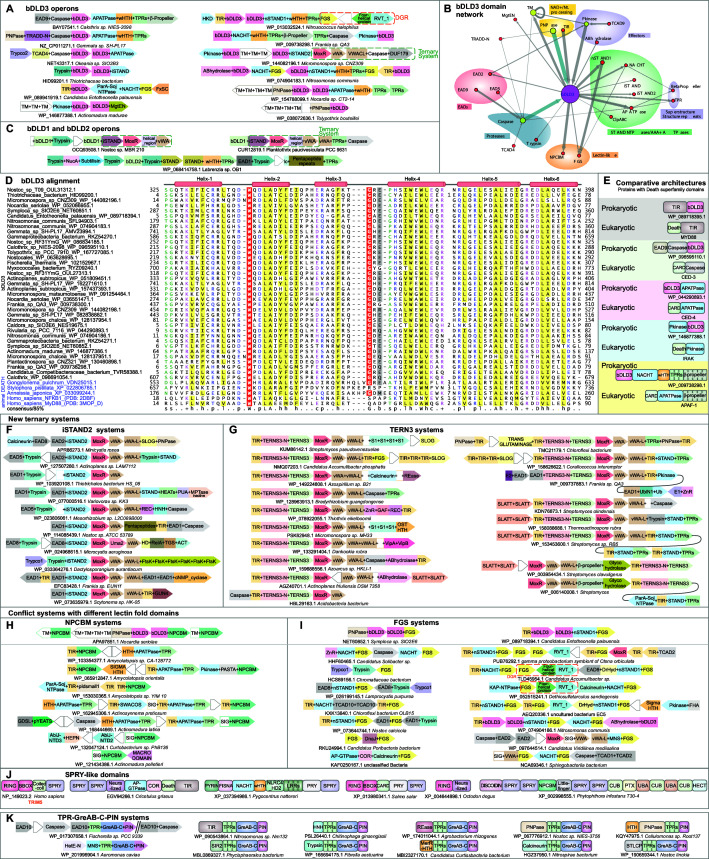
Figure 2. Domain and gene neighborhood contexts of the bacterial Death-like domains (bDLD).
(A) Representative gene neighborhoods coding for bDLD3 proteins. (B) Domains architectural network of bDLD3. (C) Representative gene neighborhoods coding for bDLD1 and bDLD2 proteins. (D) Multiple sequence alignment (MSA) of bDLD3 and representative eukaryotic Death domains (in purple). Sequences are denoted by the organism name and NCBI protein accession number separated by an underscore. Domain limits are numbered. . The predicted secondary structure and the sequence consensus at 85% identity are depicted above and below the alignment, respectively. Coloring is as per the consensus abbreviation of residue type, where s: small; u: tiny; +: basic; h: hydrophobic; l: aliphatic; p: polar; b: big. In all figures, α-helices and β-strands in MSAs are depicted as cylinders and arrows, respectively. Insertions in the sequences are represented by the number corresponding to their length. (E) Comparable domain architectures of the bDLD3 and Death-superfamily domains. iSTAND2 (F) and TERNS3 (G) containing novel ternary conflict systems. Novel conflict systems utilizing the lectin fold domains NPCBM (H) and FGS (I). (J) Domain architectures of eukaryotic SPRY-like domains. (K) Novel conflict system with a constant core comprising TPR, GreA/B-C-terminal, and PIN domains.