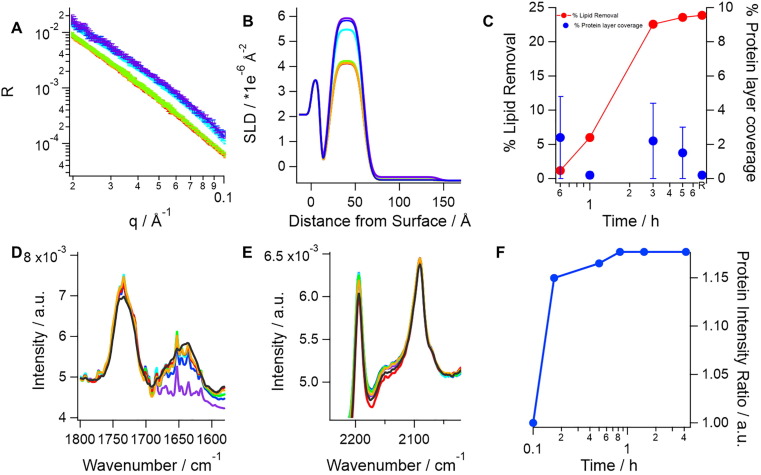
Fig. 1.
SARS-CoV-2 spike protein (0.05 mg/mL or 0,1 µM) incubation with model membranes composed of saturated lipids and cholesterol lead to lipid removal, which is accompanied by protein adsorption on top of the lipid membrane. The top panel, A shows neutron reflection profiles (markers) and best fits (lines) in h-TBS at 37 °C upon incubation time of 0 min (purple), 40 min (blue), 1 h (cyan), 3 h (green), 5 h (orange) and upon rinsing with excess h-TBS (red). Corresponding scattering length density (SLD) profiles for best fits to the neutron reflection data are shown in B. Apparent lipid removal as calculated from a change in the SLD (red) of the lipid tail layer as well as additional protein layer coverage (blue) are shown in C. Protein binding was corroborated by ATR-FTIR (bottom panel). In this case, a model membrane composed of dDMPC and hcholesterol is formed and pre-characterized in d-TBS (purple) and a broad amide I peak (~1640 cm−1, D) appears as soon as the protein is introduced in the cell (blue, 10 min; cyan, 30 min; green, 50 min; red, 90 min; black, 4 h). A small decrease in the C O stretching (D) and the CD2 symmetric and asymmetric stretch (E) bands (occurring at ~ 1734, ~2090 and ~ 2194 cm−1, respectively) suggests that at least some dDMPC was removed. The change in amide I peak intensity over time (right) shows that protein binding occurs within the first 10 min of incubation (F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)