Abstract
The core histone tails are critical in chromatin structure and signaling. Studies over the past several decades have provided a wealth of information on the histone tails and their interaction with chromatin factors. However, the conformation of the histone tails in a chromatin relevant context has remained elusive. Only recently has enough evidence emerged to start to build a structural model of the tails in the context of nucleosomes and nucleosome arrays. Here, we review these studies and propose that the histone tails adopt a fuzzy complex with DNA, characterized by robust but dynamic association. Further, we discuss how these DNA-bound conformational ensembles promote distinct chromatin structure and signaling, and that the fuzzy nature is important in transitioning between functional states.
Keywords: Nucleosome, Chromatin, Post-translational Modification, Intrinsically Disordered Regions
Histone tails: dynamic hubs for chromatin signaling
The eukaryotic genome exists in the cell nucleus as chromatin, a complex between the genomic DNA and proteins known as histones. The most basic repeating unit of chromatin is the nucleosome core particle (NCP) (see Glossary), in which ~147bp of DNA wrap around an octamer that contains two each of the core histone proteins H2A, H2B, H3, and H4 [1]. NCPs are flanked by linker DNA, which is of variable length (~10–70bp) depending on the local chromatin state [2]. The dynamic organization of these nucleosome particles, both spatially and temporally, is critical in regulation of the underlying genome and in the proper execution of all DNA templated processes [3]. Chromatin modulation is orchestrated by a slew of chromatin associated proteins (CAPs). In addition, post-translational modifications (PTMs) on the histone proteins can directly regulate chromatin or indirectly regulate it through modulation of CAP activity [4].
Much effort has been placed on building an understanding of the structure and dynamics of nucleosomes and chromatin. Several near atomic resolution structures of NCPs have been solved, and lower resolution structures of nucleosome arrays and nucleosomes in complex with CAPs are more recently being tackled. Together, these have given us great insight into chromatin structure [5–8]. In addition, mechanisms of inherent nucleosome dynamics have been characterized including DNA breathing (i.e. spontaneous reversible unwrapping) of the DNA at the entry/exit points [9–11].
However, one component has remained elusive: the histone tails. These are the N-termini of all four histone proteins, as well as the C-terminus of H2A, that protrude out from the nucleosome core. These tails are enriched in PTMs and are known to be hubs of chromatin signaling [12]. They largely do not resolve in the structures of the nucleosome or nucleosome arrays, indicating a high level of conformational dynamics, which is also demonstrated by their susceptibility to protease digestion [13]. However, a number of studies have indicated that they are not fully solvent exposed and have DNA binding potential (reviewed in [14]). The conformations of the histone tails and the mechanistic basis of their interactions with CAPs in the context of the nucleosome has thus remained elusive for a number of years. Here, we review a number of structural and biophysical studies that are beginning to shed light on histone tail conformations in the nucleosome context, and the implications in chromatin structure and signaling. We suggest that they exist in robust but dynamic complex with DNA in the chromatin context, adopting so-called “fuzzy” conformational ensembles. We further suggest that this provides an added layer of conformation-mediated regulation. Factors such as histone PTMs and nucleosome binding that modulate these conformational ensembles can directly regulate chromatin structure and indirectly regulate downstream signaling.
Studies on histone tail peptides
Each histone tail protrudes from a unique position on the nucleosome core. Though all the tails are enriched in positively charged residues, each has a unique sequence (Figure 1A), leading to distinct patterns of PTMs and interaction partners. Notably, the tails are highly conserved throughout evolution, consistent with their role in signaling.
Figure 1. Histone composition and nucleosome structure.
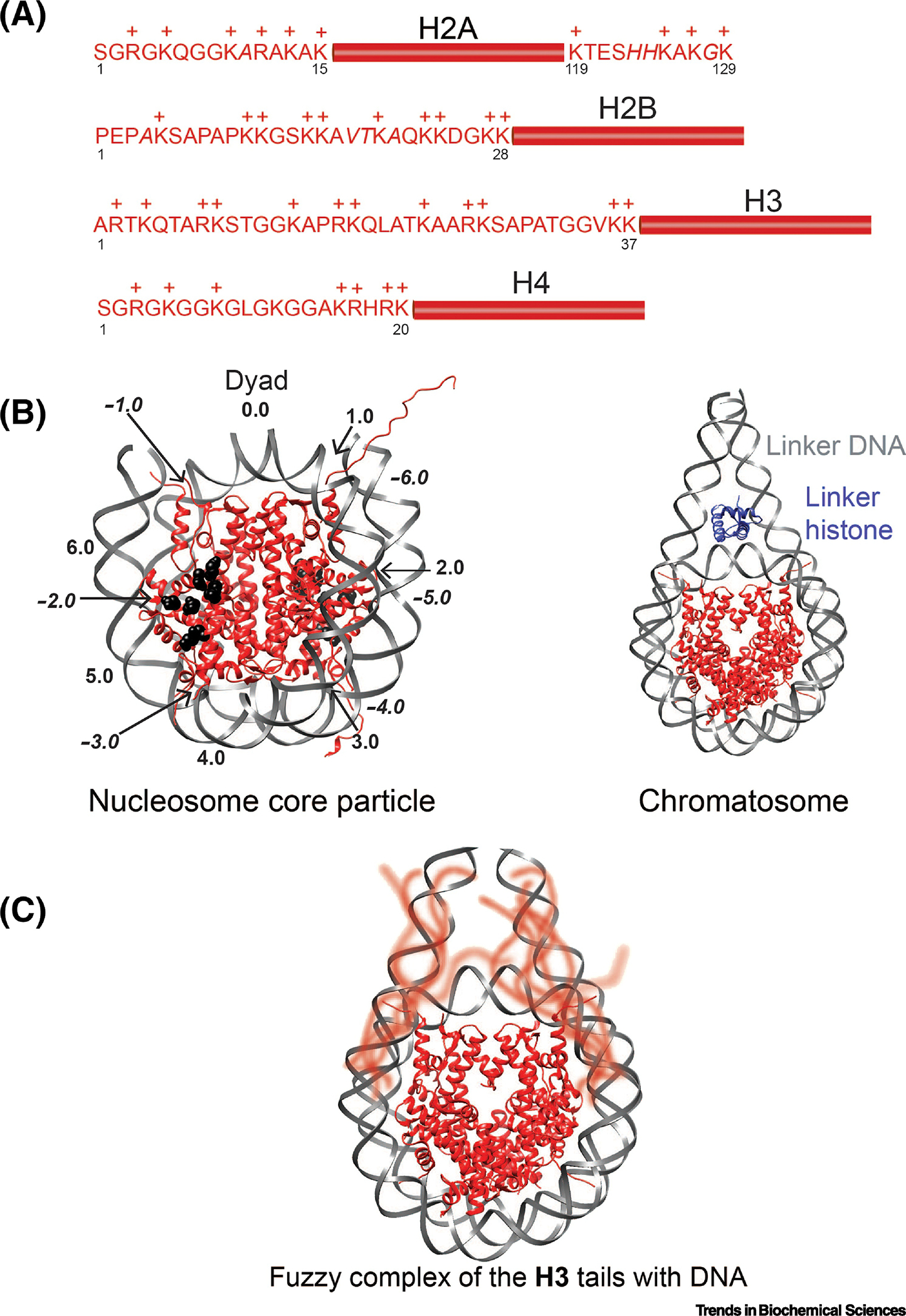
(A) Core histones H2A, H2B, H3, and H4. The core region is represented by rectangle flanked by the tail sequences. Shown are human sequences with residues that vary between organisms (only in H2A and H2B) in italics. The positive residues are denoted by a (+). (B) Left, a crystal structure of the nucleosome core particle (PDB ID 1AOI). Histones are shown in red and DNA in gray, with the H2A/H2B acidic patch residues shown as black spheres. The super helical locations (SHLs) are marked with the negative SHLs italicized. Right, a crystal structure of the chromatosome (PDB ID 5NL0). Histones are shown in red and DNA in gray, the linker histone in blue. (C) A model of the broad conformational ensemble adopted by the H3 tails in the context of the nucleosomes. The tails are blurred to represent their dynamic exchange between states.
As early crystal structures largely did not resolve the tails, many early structural studies focused on representative peptide fragments or isolated histone proteins. The conclusions from these studies were somewhat conflicting as to if the tails adopt secondary structure or not. While experimental studies largely suggested that the H2A, H2B, and H3 N-terminal tails, and H2A C-terminal tails, are primarily unstructured [15], some computational studies suggested that the H3 and H4 tails have some alpha-helical propensity [16–20].
Early studies also revealed the ability of these histone peptides to associate with DNA, which was found to be weakened upon acetylation of lysine residues [21–23]. All-atom molecular dynamics (MD) simulations suggested that the charge neutralization upon DNA binding can increase the helical content of the H3 tail [19]. Thus, even these early studies indicated that the tail DNA contacts could be important in structure and signaling. However, their structural status in the context of the nucleosome remained unclear.
Histone tail conformations in the context of the nucleosome
While studies on peptide fragments of the histone tails are informative, they do not take into account the context of the nucleosome. While each tail has a unique conserved sequence (Figure 1A), within the nucleosome context, the tails are further distinguished by their unique positioning with respect to the DNA wrap and core fold of the histone proteins. Lack of resolution of the tails in crystal structures of NCPs, and susceptibility to digestion both indicate a high degree of conformational flexibility of the tails [13,24]. This often leads to depictions of the tails as being extended away from the nucleosome core and fully accessible to modification and CAP association. However, biophysical and biochemical studies have indicated that the tails do have some effect on the nucleosome core, and it was hypothesized that this may be due to direct interactions. These earlier studies have been reviewed previously [5,14,25,26]. Below, we focus on the most recent findings for each of the histone tails. These utilize a variety of structural and biophysical approaches (Box 1) that together help paint a picture of their conformations in the nucleosome context. Note that the term nucleosome will be used to refer to the NCP or NCP containing additional linker DNA. Where important this will be distinguished.
Box 1: Structural and biophysical methods for studying histone tails.
Characterizing the conformational ensemble and dynamics of the histone tails in the nucleosomal context has been challenging as they exchange between a broad range of conformations. Furthermore, reconstitution of nucleosomes in large quantities can be challenging, especially if specific post-translational modifications (PTMs) are desired [100]. Outlined below are methods that have been especially useful for studying histone tails conformation and dynamics.
Hydrogen /Deuterium Exchange Mass Spectroscopy (HDX-MS) measures the exchange of protein backbone amide protons with solvent deuterium over time. The rate of deuterium exchange depends on the amide accessibility to solvent. This technique can quantify the exchange rates to single-residue resolution if coupled with proteolytic digestion. It can be applied to conformationally heterogenous samples, does not have a size limitation and consumes small amounts of sample (< 0.1 mg).
Nuclear Magnetic Resonance (NMR) spectroscopy takes advantage of the magnetic properties of nuclei providing atomic level information on the structure, dynamics, and interaction of molecules. NMR is ideal for investigating the structure of conformationally dynamic and disordered proteins. It can provide structural information as well as dynamical information from pico-second to second timescales.
Forster Resonance Energy Transfer (FRET) takes advantage of the non-radiative transfer of excitation photon energy from one fluorescent dye (donor) to another dye (acceptor) providing distance information in biomolecules. It can be applied to heterogenous samples, is able to monitor conformational dynamics with time resolution from nano-second to minutes or longer. In addition, single-molecule FRET (smFRET) can provide information about transient intermediates of a pathway and about the heterogeneity among different molecules that is undetectable in bulk measurements. The drawbacks are that the sample must be properly engineered with dyes and it is often insensitive to small amplitude motions.
Molecular Dynamics (MD) simulations is an in silico approach to investigating conformational dynamics, where molecular motions are calculated as a function of time. Depending on the approach, it can provide atomic level detail of sub micro-second motions, to lower resolution on longer timescales.
Histone H3 tails
The H3 N-terminal tails are positioned one each on either side of the nucleosome dyad near the entry and exit DNA. They extend through the DNA gyres near DNA super helical locations (SHLs) +6.5/−0.5 and −6.5/+0.5 (Figure 1B). Based on crystal structures, residues 1–37 exist outside of the core and usually do not resolve, and residues 1–27 are sensitive to trypsin digestion [13]. The H3 tails appear to be largely intrinsically disordered in the context of the nucleosome with or without linker DNA, with only moderate helical propensity observed. While circular dichroism studies have suggested the H3 tails adopt up to 50% helical content [27], NMR and all-atom MD simulation studies indicate that the unmodified H3 tail is largely intrinsically disordered [28–30]. However, the helical content can be regulated by post-translational modification, increasing upon acetylation of lysines [17,31].
The H3 tails have not been observed to have a large effect on the overall structure of nucleosome core [32–34], but do impact its conformational dynamics, in particular the breathing motions of the DNA. Specifically, truncation of the first 27 residues of the H3 tail in nucleosomes with or without linker DNA leads to differences in DNA breathing dynamics, with an observed increase in the population of the open state [35,36]. Consistent with this, truncation of H3 has previously been found to have a small but detectable decrease in the stability of the nucleosome core [37–40]. This effect is also observed in computational studies of the nucleosome. Though shorter simulations do not capture DNA breathing dynamics, spontaneous unwrapping of the entry/exit DNA is observed in long (10s of microseconds) or high-temperature all-atom simulations, or in coarse-grained simulations. In these studies, truncation of the H3 tail on a nucleosome with or without linker DNA is observed to affect the DNA breathing dynamics, changing the overall conformational ensemble and rate of unwrapping [41–43]. In one coarse-grained simulation of a nucleosome with linker DNA, the H3 tail and DNA dynamics were observed to be correlated [42]. Notably, these effects have only been explored for a very limited number of DNA sequences that form stable nucleosome core structures in vitro, and effects could be even larger for different sequences.
Despite the apparent conformational heterogeneity of the H3 tails, they are not seen to be extended, but rather have been found to be largely collapsed onto the nucleosome with or without linker DNA. Native mass spectrometry (MS) indicate that the H3 tails adopt a primarily compact conformation [44], and hydrogen-deuterium exchange MS (HDX-MS) support that they are protected from exchange in the context of the nucleosome [45]. This collapsed conformation is consistent with observations of direct interaction of the histone H3 tails with the nucleosome core with or without linker DNA [28,30,46]. NMR, cross-linking, and a variety of MD simulations all support that the entire H3 tail makes contacts with nucleosomal and/or linker DNA [28,30,31,42,43,47,48]. This is seen to primarily be driven through contacts with lysine and arginine, but also includes contributions from polar residues in the tail [30]. Importantly, these contacts are seen even at physiological salt concentrations. Consistent with the H3 tails having broad conformational heterogeneity, the H3 tails are seen to adopt multiple conformations on nucleosomal DNA in multiple all-atom MD simulations [30]. In addition, cross-linking studies with nucleosomes containing linker DNA reveal contacts with the DNA at several positions including with both the nucleosomal DNA and linker DNA [48]. Notably, the points of contact with the nucleosomal DNA (H3T6 with SHL +/−1.5 and +/2.0, and H3A15 with SHL +/−0.5, +/−6.5, and +/−7.0) are primarily around the entry/exit DNA which is consistent with the H3 tails having an effect on DNA breathing dynamics.
Despite extensive interactions with nucleosomal and linker DNA, the H3 tails undergo a high level of conformational dynamics. These dynamics appear to span a broad range of time-scales, from nano-second to second. NMR analysis indicates that the tails are dynamic on the sub-microsecond timescale along the entire length of both tails [30]. Additionally, a single molecule FRET study found that the tails exist in at least four main configurations, three with DNA near the dyad or linker DNA, and one with the histone core [46], with transitions between configurations taking place on micro- to milli-second time-scales. Finally, HDX-MS support exchange on the time-scale of seconds, with the exchange rate decreasing from the N-terminal to C-terminal end of the tail [45]. Together, this suggests a model in which the H3 tails forms robust interactions with nucleosomal and/or linker DNA on the macroscopic level, while microscopically undergoing fast exchange between states of a large conformational ensemble (Figure 1C) [30].
Histone H4 tails
In contrast to the H3 tails, which extend through the DNA gyres, the H4 N-terminal tails extend from the face of the nucleosome. Specifically, they are positioned one each on either face of the nucleosome, extending from the core near SHL +/−2.0 (Figure 1B). Based on the crystal structure, the N-terminal alpha-helix of H4 ends at residue 23, though the tail is only sensitive to trypsin digestion through residue 20 [13]. Like H3 tails, the H4 tails are not extended but collapsed onto the nucleosome core, with HDX-MS supporting a primarily compact conformation [45]. Cross-linking also supports that the H4 tails interact with DNA, however, in contrast to the H3 tails, these contacts are seen entirely with nucleosomal DNA (H4L10 with SHL +/−1.5 and +/−2.5) [48]. Removing the H4 tails does not have a significant effect on DNA breathing dynamics. However, removing the H4 tails in addition to the H3 tails leads to a substantial increase in the DNA entry/exit end to end distance as compared to removing the H3 tails alone [35]. This suggests cross-talk between the H3 and H4 tails, which may be due to altered conformational ensembles of the DNA bound state of one tail upon perturbation of the other (Box 2).
Box 2: Histone tail ‘crosstalk’.
Histone post-translational modifications (PTMs) are thought to function in combinations, with certain PTM patterns contributing to a downstream effect [101,102]. Mechanisms of cis-tail crosstalk have been studied at the level of peptides and revealed that single or multiple reader domains can recognize (or exclude) multiple PTMs. However, we know very little about these mechanisms in the context of the nucleosome. Consideration of a DNA-bound histone tail suggests a new avenue for cis-tail PTM crosstalk (Figure 2). Namely, PTMs that alter the conformational ensemble of a tail could alter its accessibility and thus indirectly regulate binding. Consistent with this, phosphorylation of the H3 tail in the context of the nucleosome was observed to indirectly enhance BPTF PHD finger binding to H3K4me3 as well as promote lysine acetylation [28,30]. Conversely, reducing the conformational ensemble would further restrict tail binding. Though this has not yet been observed for any PTMs, linker histone H1 has been reported to reduce H3 tail conformational dynamics and subsequent acetylation and phosphorylation of the tail [28].
Trans-tail PTM crosstalk was first revealed between yeast H2BK123 ubiquitylation and H3K4 methylation [103,104]. This type of crosstalk has since been observed for other PTM combinations including H3K79 methylation and H2BK123 (mammalian K120) ubiquitylation, H2BK34 ubiquitylation and H3K4 and H3K79 methylation, and H4K20 methylation and H3K9 methylation [105–107]. The molecular mechanisms of this crosstalk have been partially elucidated and include direct effects on enzyme recruitment, positioning, and activity [108]. However, the conformation of the histone tails may also be contributing to this crosstalk in an indirect way. This could occur through localized effects, where changes to the conformational ensemble of one tail may alter an adjacent tail, likely through altering the available DNA surface. Indeed, this mechanism has been proposed to be underlying the regulation of the H2B/H3 crosstalk by the H2A N-terminal tail [109]. Specifically, it was proposed that changes to the H2A tail/DNA interaction alters the modification of H2B and subsequently H3. Similarly, acetylation of the H4 tail was recently found to affect the dynamics of the H3 tail [110]. Trans-tail crosstalk could also propagate through allosteric networks across the nucleosome core. These types of networks have been identified in molecular dynamics (MD) simulations and, notably, were altered upon incorporation of histone variants [111]. Allosteric communication between the α3 helix of H2A and the H3 N-terminal tails has also been observed [46,112,113]. Finally, loss of the H2A/H2B dimer has recently been observed to alter the conformational dynamics of the directly adjacent H3 tail, and also alters the conformational ensemble of the non-adjacent H3 tail, proposed to be induced by changes in the DNA dynamics [76]. Thus, the conformation of the tails in the context of the nucleosome is likely a critical contributor to chromatin PTM crosstalk.
Beyond positioning, a major difference between the H3 and H4 tails is in the conformational dynamics. Whereas the H3 tail appears to have similar dynamics along its full sequence, the H4 tail contains two distinct dynamical regions. This difference is likely due both to positioning of the tails, as well as sequence differences. In particular, H4 residues 16–23 contain a high density of arginine and lysines referred to as the H4 basic patch that is positioned close to the nucleosome core (Figure 1A). NMR analysis of the H4 tail in the context of the nucleosome indicates that the first 15 residues undergo fast (sub-microsecond timescale) conformational dynamics, while residues 16–23 undergo slower (micro-seconds or slower) dynamics [29]. Similarly, HDX-MS indicates a decrease in the level of deuterium uptake when comparing the first 15 residues of the tail to residues downstream of Lys16 [49].
The slow conformational dynamics of the H4 tail basic patch is consistent with crystal structures in which these residues are often resolved and seen to form intra- and/or inter-nucleosome interactions with DNA or the H2A/H2B acidic patch in the crystal lattice. Notably, the exact conformation is heterogeneous from structure to structure, indicating a range of possible configurations. While several studies have confirmed an interaction between the H4 tail with an adjacent H2A/H2B acidic patch in a nucleosome array [50,51], recent cross-linking studies reveal that the H4 tail also forms intra-nucleosome interactions with the H2A/H2B acidic patch and DNA [52–54].
Histone H2A tail
H2A has both N-terminal and C-terminal tails. Similar to the H4 tails, the H2A N-terminal tails extend from each face of the nucleosome, but are further from the dyad than the H4 tails, near SHL +/−4.5 (Figure 1B). The first 12–15 residues are generally not resolved in crystal structures and residues 1–12 are sensitive to trypsin digestion [13]. In comparison, the C-terminal tails of H2A are positioned very close to the dyad (SHL +/− 0.5) near the H3 tails. They are sensitive to digestion from residue 119 to the C-terminus [13]. Cross-linking studies indicate the H2A N-terminal tails interact with nucleosomal DNA near SHL +/−4.0 [55,56]. The C-terminal tails interact with nucleosomal DNA as well as linker DNA, similar to the H3 tails. All-atom MD simulations and crosslinking studies on nucleosomes with linkder DNA indicate that the nucleosomal contacts occur near the dyad and at SHL +/−6.0 [43,55–58], and are modulated by binding of linker histone [56]. Similar to the H3 tails the H2A C-terminals tails were suggested to stabilize DNA wrapping [47,58].
NMR studies indicate that residues 1–10 in the N-terminal tails and 118–123 in the C-terminal tails are unstructured and dynamic on the fast (sub-microsecond) timescale [29], whereas residues 11–15 in the N-terminal tail were not visible indicating slower dynamics. Together, this suggests that the H2A C-terminal tail conformational dynamics are similar to the H3 tail, experiencing fast conformational dynamics along the length of the tail. In contrast, the H2A N-terminal tail is more comparable to the H4 tail, with the N-terminal residues undergoing faster dynamics as compared to the residues closest to the core, which is consistent with the latter resolving in some crystal structures.
Histone H2B tail
Similar to the H3 N-terminal tails, the H2B N-terminal tails protrude between two DNA gyres. However, in contrast to the H3 tails, they are positioned away from the dyad near SHL +4.5/−2.5 and −4.5/+2.5 (Figure 1B). This is very close to where the H2A N-terminal tails are positioned. Residues 1–28 of the H2B tails usually do not resolve in crystal structures and 1–24 are sensitive to trypsin digestion [13]. As with the H3, H4, and H2A tails, the H2B tails have been shown to interact with DNA, making contacts around SHL +/−5.0 and SHL +/−3.0 [58,59]. A chemical reactivity assay indicates that the entirety of the tail interacts with DNA, but, reminiscent of the H4 and H2A N-terminal tails, there appear to be at least two elements along the tail with distinct affinity for DNA. Specifically, the N-terminal portion of the tail binds weaker than the C-terminal portion. It is not yet clear if this pattern of dynamics is due to positioning or sequence of the tail, though notably there is a basic patch at the C-terminus reminiscent of the H4 tail basic patch (Figure 1A).
Consistent with the distinct regions of affinity, distinct regions of dynamics are observed in all-atom and targeted MD simulations [58]. The N-terminal residues (1–23) sample a broad range of heterogenous conformations on the DNA surface, while the C-terminal residues adopt a more restricted conformational state in the DNA minor groove [58]. Consistent with this NMR spectroscopy studies indicate that only residues 1–24 undergo fast conformational dynamics (sub-microsecond) on the NMR timescale [29]. Interestingly, while deletion of the H2B tails has been found to decrease the stability of the nucleosome overall [34], it has also been found H2B tail deletion decreases the mobility of the histone core on DNA [40,58]. This effect was enhanced by deletion of the H4 tail, again indicating trans-tail cross-talk (Box 2). It has been proposed that the interaction of the N-terminal residues of the H2B tail with DNA stabilize the nucleosome, while the C-terminal residues actually destabilize the nucleosome, promoting looping of the DNA [58].
Together, these studies on the core histone tails indicate that, in the context of the nucleosome, all of the tails are in a primarily DNA-bound state. Collectively they cover the entire DNA wrap (i.e. make contacts at every SHL), as well as make contacts with linker DNA. While these contacts do not appear to alter the structure of the core, they alter the stability, and the tails positioned near the entry/exit DNA alter the DNA breathing dynamics. In addition, this presents a major barrier to DNA binding. The tails do not adopt static conformations but rather experience a high level of conformational dynamics in the DNA-bound state (Figure 1C). Interestingly, along a single tail there may be different dynamical regions, all of the tails sample a range of conformations with fast transitions between multiple states. This is consistent with a so-called fuzzy complex (Box 3). As noted in the next sections, tail/DNA interactions appear to be critical in chromatin signaling and regulation, and we propose that their fuzzy nature is important in transitioning between functional states.
Box 3. Histone tails adopt “fuzzy” complexes with DNA.
As outlined above, it is clear from biophysical, biochemical, and structural studies that the histone tails are largely intrinsically disordered and highly conformationally dynamic. However, a number of biochemical, and more recently structural, studies have supported that they associate with nucleosomal and linker DNA. Moreover, these are not transient weak interactions, but rather appear to be robust in nature, capable of competing with histone binding domains. Together, this indicates that the histone tails in the context of chromatin form what are known as “fuzzy” complexes with DNA. First introduced in 2008 by Tompa and Fuxreiter [96], the term “fuzzy” in relation to protein/protein interactions was coined to define complexes in which a substantial amount of disorder existed in the bound state, that could not be ascribed to a single conformation or even small number of well-defined conformations. Rather, the bound state is defined by a large conformational ensemble with small energy barriers between states [98]. This is favorable due to a reduction of the entropic penalty upon binding (in this case DNA). In addition, as both histone tails and DNA are polyelectrolytes, this is also entropically favorable due to large solvent release. Despite the dynamic nature of these complexes, they can be high-affinity [114,115]. We posit that, similarly, the tails form high affinity interactions with DNA while retaining a high level of disorder. These ensembles are likely characterized by fast microscopic dissociation of small regions of each tail facilitating transition between conformational states, while maintaining slow macroscopic dissociation of the whole tail, leading to overall high affinity. Each tail likely samples different ensembles based on the exact amino acid composition (including distribution of charged residues) and orientation within the nucleosome. Notably, conformational freedom will also be limited by the fact that it is tethered to the nucleosome core. Importantly, these bound state ensembles will be modulated through post-translational modification of the tail, making them critical in chromatin signaling.
The effect of histone tail conformation on chromatin signaling and regulation
A major functional role of the histone tails is in chromatin signaling, with the tails experiencing a high level of PTMs. These PTMs are recognized by histone reader domains, mediating the interaction of various CAPs with modified nucleosomes and regulating CAP activity. Until recently, studies of histone tail binding have been largely carried out using peptide fragments of the histone tails [12,60]. However, this neglects the effect of tail/DNA interactions. In fact, for several reader domains, histone tail binding affinity has been found to be substantially reduced in the context of the nucleosome [30,61,62]. While multivalent contacts can partially overcome this, a decrease in affinity is observed even for multiple domains [62]. This supports that tail/DNA interactions are robust and have a dramatic effect on chromatin signaling [28,30,61,62]. Notably, this effect is not limited to reader domains alone. Several histone modifying proteins or complexes have been found to be inhibited in the nucleosome context, as well [28,63].
Importantly, this gives rise to mechanisms of PTM cross-talk. Histone PTMs often act in combination, which is usually thought to involve direct recognition of multiple PTMs. However, strong interaction of the histone tails with DNA implies that within a specific functional pattern of PTMs, some may not be directly recognized but rather be functioning to alter histone tail dynamics (Figure 2). Indeed, charge-modulating PTMs have been found to alter the conformational dynamics of the tails on DNA. For example, serine/threonine phosphorylation and lysine acetylation of the H3 tail have been found to weaken the interaction with both nucleosomal and linker DNA and promote tail accessibility to readers and modifying enzymes [28,30,46]. Similarly, mutation of H4K16 to Q increases the dynamics of the H4 tail basic patch, and mutation or acetylation of lysine residues in the H2B tail results in tail compaction and weakening of the interaction with DNA [59, 61]. Notably, these histone PTMs do not abrogate the interaction of tails with DNA, but rather modulate the affinity and conformational ensemble and dynamics of the DNA-bound state. Recent evidence suggests that PTMs may not only modulate tails in-cis but could affect tails in-trans (Box 3). Importantly, the effect of only a few PTMs have been tested to date compared to the plethora of known histone PTMs (Figure 3). The diversity of these PTMs will likely lead substantial variances in the magnitude and nature of the effect on tail/DNA interactions.
Figure 2. Schematic model of histone tail contacts in various chromatin states and regulatory effects of tail/DNA interactions.
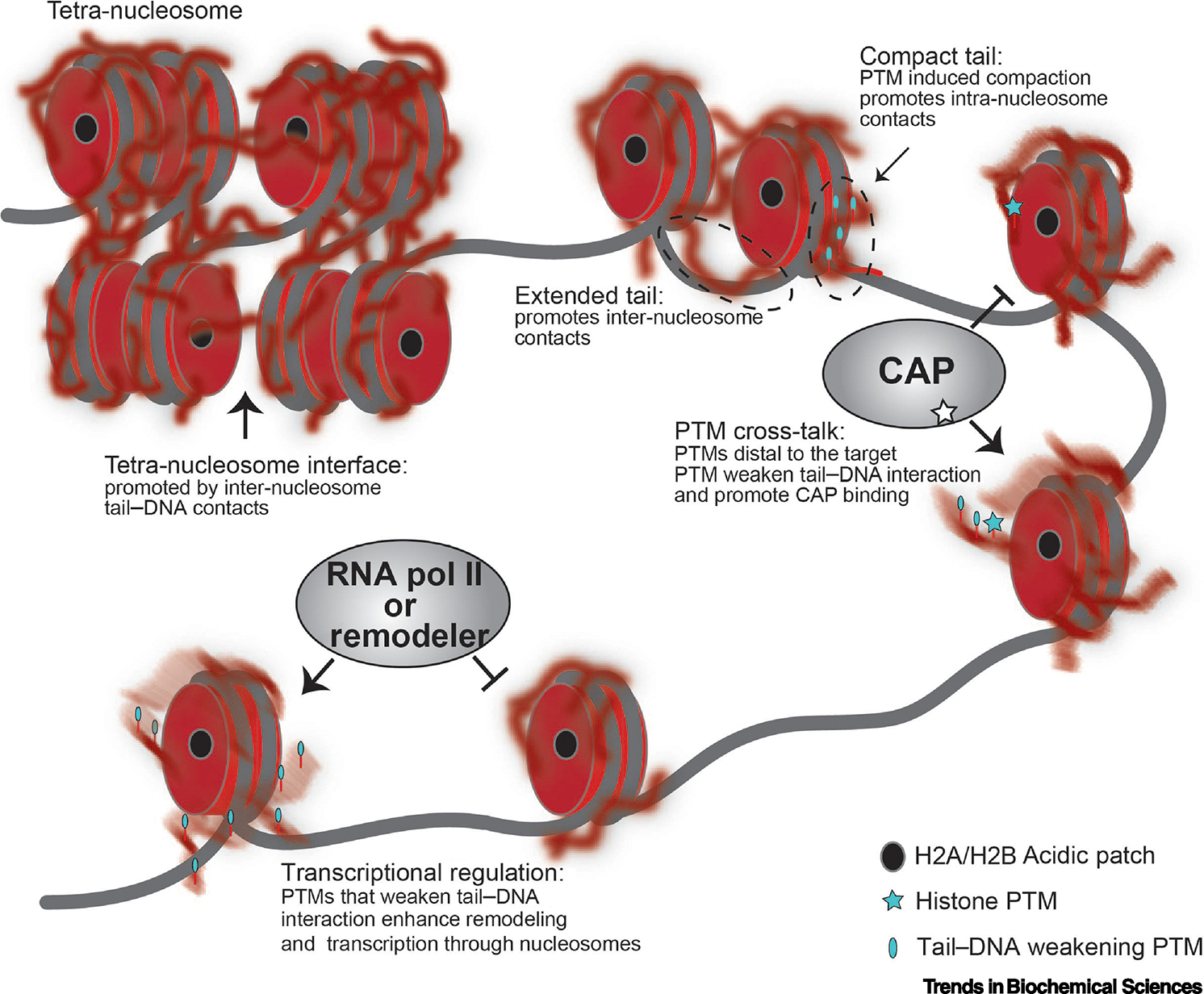
Histones are shown in red, DNA in gray, the H2A/H2B acidic patch as a black oval, and histone post-translational modifications (PTMs) as cyan ovals or stars. The histone tails are blurred to represent dynamic exchange within a broad conformational ensemble. Predicted inter-nucleosome interactions stabilizing the tetra-nucleosome are shown. The intra- and inter-nucleosome interactions favored by compact and extended tails are shown. A chromatin associated protein (CAP) is shown with a star denoting a histone PTM binding pocket, and the effect of PTM crosstalk on CAP binding is represented. The inhibitory effects of RNA polymerase II (RNA Pol II) and remodeler activity, as well as the positive effect of tail/DNA weakening PTMs, are represented.
Figure 3. Histone modifications.
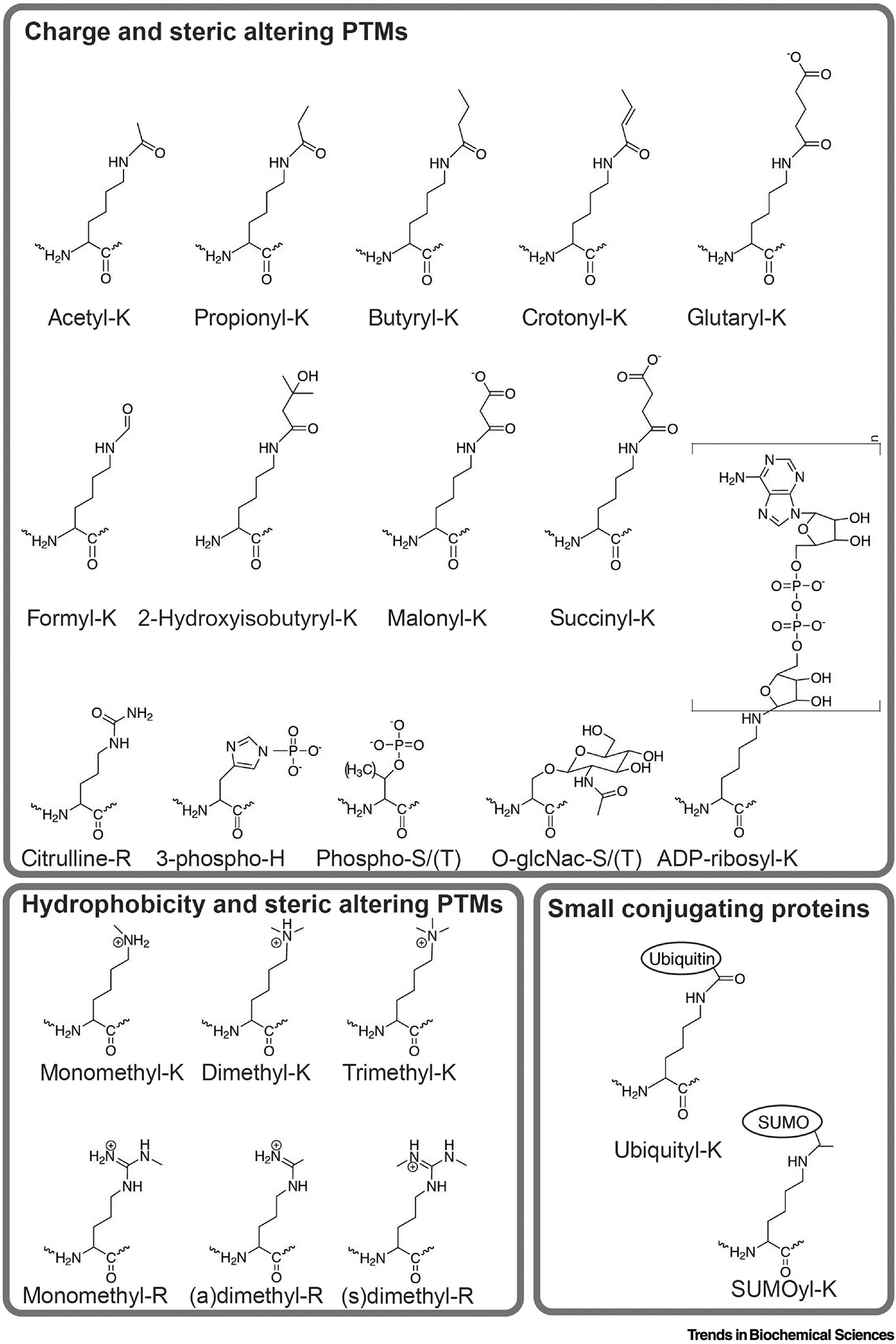
Shown are chemical structures for various histone modifications that are expected to change charge/sterics (top), hydrophobicity/sterics (bottom, left), or all upon conjugation of small proteins such as ubiquitin or SUMO (bottom, right). These are all expected to alter tail/DNA interactions.
In addition to regulation by PTMs, tail/DNA interactions can be modulated by DNA binding factors. Several reader domains have been found to bind to DNA in addition to the histone tail (see [64] and references therein). For these domains, affinity is actually higher for the nucleosome as compared to histone peptides. Importantly, beyond providing multivalent contacts, it has been suggested that reader domain interaction with DNA may also help displace the histone tails from DNA to expose them for binding [30,64,65]. In some cases, auxiliary DNA binding domains or proteins may play a similar role [62,66]. For instance, HMGN1/2 binding to the nucleosome has recently been shown to alter the conformational ensemble of the H3 and H4 tails on DNA [48]. Notably, not all DNA-binding factors lead to increased accessibility of the tails. Binding of the linker histone H1 to form the chromatosome (Figure 1B) was found to reduce H3 tail dynamics and accessibility potentially stabilizing it on the linker DNA [28]. Similarly, a recent study found that the H3 tail stabilizes an RNA/DNA triplex formed on the linker DNA of a nucleosome [67]. Thus, the accessibility can be even more restricted by limiting the available conformational ensemble.
The weakening of tail/DNA interactions can also regulate machinery acting on the nucleosome core independent of tail binding (Figure 2). For instance, the presence of histone tails decreases progression of RNA polymerase II through a nucleosome. However, mutation of lysine residues to mimic acetylation in the tails, which would weaken tail-DNA interactions, positively regulates RNA polymerase II activity, enhancing progression [68,69]. Similar effects are seen with chromatin remodelers. Acetylation of the H3 tails increases the affinity and activity of SWI/SNF [70]. Similarly, deletion of the H2A C-terminal tails increases the activity the ISWI remodelers [71]. For INO80, deletion of the histone tails leads to conformational heterogeneity of the nucleosome upon initiation of remodeling [72] and faster remodeling activity [73], indicating that the tails restrain the conformation of the INO80 bound state and remodeling activity. This regulation is likely due to multiple effects including enhanced accessibility to binding the core DNA, increased dynamics of the DNA, and increased mobility of the histone core.
Correlation of H3 and H2A C-terminal tails with DNA breathing dynamics presents a number of avenues for nucleosome regulation. For instance, the binding of the PHF1 Tudor domain or CHD3 PHD finger reader domains to the H3 tails promotes partial unwrapping of the DNA [74,75], indicating that H3 tail binding is likely to regulate DNA binding dynamics. Inversely, unwrapping of the DNA has been shown to alter the H3 tail dynamics and accessibility. Specifically, loss of an H2A/H2B dimer leads to an increase in the adjacent H3 tail dynamics [76]. This was proposed to be due to a decrease in DNA density with unwrapping of the DNA concurrent with the loss of dimer. For factors targeting partially unwrapped nucleosomes or subnucleosomal particles as well as the H3 tails, this effect could lead to cooperative binding and substantially increased activity.
Histone tails in nucleosome arrays and higher order chromatin structure
Beyond the localized interactions of the tails with nucleosomes and effects on chromatin signaling and regulation, the histone tails have long been implicated in the assembly of higher-order chromatin structures. Several studies have been performed to assess the effect of histone tails on inter-nucleosome interactions, nucleosome array compaction, and array oligomerization, which have been reviewed previously [26,77]. Importantly, the mechanism by which the tails contribute to these interactions is not due solely to charge-neutralization and reduction of inter-nucleosome repulsion [78–80], but rather the tails have been shown to form critical inter-nucleosome interactions [81]. Overall, the order of contribution of histone tails to compaction is H4 > H3 > (H2A/H2B), though the tails are thought to act cooperatively [14,77,78]. The conformational details of the inter-nucleosome interactions are still being elucidated. However, it is becoming clear that contacts with DNA are key in the formation and regulation of these structures.
Several experimental and computational studies have revealed that an inter-nucleosome interaction between the H4 tail basic patch and H2A/H2B acidic patch is important in compaction of nucleosome arrays [6,7,24,82,83]. In particular, H4K16 is critical and acetylation of this residue inhibits nucleosome array folding as much as deletion of the entire H4 tail [51]. Notably, modification of the H4 tail basic patch can also stabilize a condensed chromatin state, as a methyl-lysine analogue at H4K20 has been observed to enhance the folding of nucleosome arrays [84]. In addition to interactions with the acidic patch, inter-nucleosome contacts of the H4 tail with DNA are also important [53]. Interestingly, a crystal structure of a tetra-nucleosome and cryo-EM reconstruction of a 12-mer array indicate that the H4 tail interaction with the acidic patch does not occur between every nucleosome in the tetra-nucleosome, but rather at the interface between tetra-nucleosome subunits [6,7], potentially explaining the importance of the DNA contacts.
Inter-nucleosome contacts between the H3 tail and DNA are also important for compaction of nucleosome arrays [51,76] and array oligomerization [53,85]. In contrast to the H4 tail, which makes inter-nucleosome contacts along the entire tail, inter-nucleosome contacts were primarily seen with the N-terminal portion of the H3 tail [53]. H3 tail inter-nucleosome contacts were also found to be important both within the tetra-nucleosome and in oligomerization of tetra-nucleosomes into arrays [86]. Notably, a solid-state NMR study carried out on a 17-mer nucleosome array indicated that H3 residues 1–35 and H4 residues 1–21 remain very dynamic even in a highly compacted array, suggesting that they sample a broad conformational ensemble in inter-nucleosome states similar to intra-nucleosome states [87].
Several recent MD simulations (including all-atom, coarse-grained, and steered) have investigated mechanisms of inter-nucleosome contacts between mono-nucleosomes or within nucleosome arrays. In agreement with cross-linking studies, these suggest that all of the tails form inter-nucleosome contacts primarily through interaction with DNA [88–91]. These include contacts with linker DNA and nucleosomal DNA of an adjacent nucleosome. Interestingly, which histone tail dominates these interactions is seen to be dependent on nucleosome spacing and the concentration of linker histones present [88]. Recently, tail/DNA contacts have also been implicated in liquid-liquid phase separation of reconstituted nucleosome arrays, which are seen to form tail dependent droplets [92], and are proposed to be critical in mediating higher order chromatin organization.
Similar to intra-nucleosome interactions, inter-nucleosome tail/DNA interactions have been shown to be modulated by acetylation of lysine [26]. Modification of the tails appears to alter the conformational ensembles of the DNA-bound state. Specifically, computational studies suggest that acetylation does not abrogate DNA interactions, but rather leads to a more compact conformational ensemble of the tails shifting them away from inter-nucleosomal interactions and promoting intra-nucleosomal interactions [93,94]. This is also consistent with observations regarding phase separation, in which the density and stability of the droplets are reduced upon histone tail acetylation [92].
The functional benefits of a fuzzy complex
A major benefit of intrinsically disordered regions (IDRs) is their malleability, enabling a diversity of interaction partners [95]. As outlined above, the histone tails interact with a broad diversity of partners, from multiple DNA elements to a variety of CAPs. Upon association with their binding partners, IDRs can adopt secondary structure, remain unstructured but adopt a single conformation, or remain disordered and conformationally dynamic. The latter is referred to as a fuzzy complex [96–98] (Box 3). We have suggested that all of the histone tails adopt a fuzzy complex with DNA in the context of chromatin, which regulates chromatin structure, CAP activity, and access to the underlying DNA. The major functional benefit of adopting a fuzzy complex is the relative ease of regulation of the DNA-bound state [96]. This is due to the fast local dissociation of small regions of the histone tails. This allows for greater accessibility to post-translational modification. In addition, this partial dissociation can facilitate quick searching and transfer to a new binding partner as compared to a rigid bound state. This would facilitate quick transitions from inter- to intra-nucleosome interactions or vice versa in the regulation of higher order chromatin structure. In addition, it would facilitate the quick modification of the tails and interaction with CAPs. Finally, this dynamic state will transiently expose different DNA regions on the nucleosome itself, facilitating the interaction of transcriptional machinery and chromatin remodelers with nucleosomal and linker DNA.
Concluding Remarks
Altogether, these studies suggest that all of the histone tails are DNA bound in the chromatin context. It appears that no matter the state, the tails are conformationally dynamic and exchange between multiple DNA-bound conformations. However, unique DNA-bound conformational ensembles contribute to unique chromatin states (Figure 2). The observed transitions between states are quite fast (nano-second to milli-second), though this does not preclude that there are also slower transitions occurring. The interaction with DNA appears to be robust as it can substantially inhibit binding of additional factors. Modulation of these conformational ensembles are critical in chromatin structure remodeling and signaling. Factors including nucleosomes spacing, histone PTM, and chromatin binding proteins have the potential to shift these ensembles and thus regulate the chromatin state. Though structural models are emerging, there is much yet to be determined about the histone tail conformations, dynamics, and modulation (see Outstanding Questions). Importantly, there is also a dearth of in vivo data addressing the interaction of the histone tails with DNA. Though, Rhee et al. have identified H3 tail interactions with linker DNA in cells [99], there is much to be learned about the impact of tail-DNA interactions on cellular processes.
Outstanding Questions Box.
Though a few studies have investigated the effect of lysine acetylation or serine/threonine phosphorylation on tail/DNA interactions, a large number of post-translational modifications (PTMs) remain unexplored. How do other histone PTMs alter tail/DNA conformational ensembles and dynamics?
What is the conformational ensemble and dynamics of the tails of histone variants?
How do the tails affect the RNA/chromatin interface and inversely how does RNA effect tail/DNA interactions?
How do disease mutations in histones (such as the onco-mutations identified in the H3 tail) alter tail/DNA interactions? What is the impact of this on chromatin associated protein (CAP) function?
The molecular details of tail/DNA interactions are becoming clearer at the single nucleosome level. In nucleosome arrays what are the molecular determinants of tail conformations?
What is the importance of these interactions in a cellular context? How do the in vitro generated models translate to in an in vivo context?
Highlights.
Eukaryotic DNA is wrapped around histone proteins to form nucleosomes that fold into higher-order chromatin structures, and the local chromatin structure regulates all DNA-templated processes
All core histone proteins contain intrinsically disordered “tail” regions that protrude from the DNA-wrapped core and are known to be critical in chromatin regulation
Recent studies have revealed that the core histone tails adopt multiple conformations on the nucleosomal and linker DNA; these tail/DNA interactions are robust, but exchange quickly between multiple conformations consistent with a so-called “fuzzy” complex
Intra- versus inter-nucleosome contacts by the tails differentially contribute to the local chromatin state and thus regulation of DNA-templated processes
Histone post-translational modifications and chromatin associated factors can modulate these fuzzy conformational ensembles and tail accessibility, indicating that the tail/DNA interactions are an important regulatory mechanism of chromatin
Acknowledgements:
HAF was supported by an NIH T32 fellowship (2T32GM008365-26A1) through the Center for Biocatalysis and Bioprocessing. The Musselman lab is funded by the National Institutes of Health (R35GM128705).
Glossary
- Chromatin-associated proteins (CAPs)
all ‘non’-histone proteins that are frequently or transiently bound to and/or processing chromatin
- Chromatosome
a nucleosome in complex with linker histone
- DNA breathing
the spontaneous reversible unwrapping of the octamer by the nucleosomal DNA at the entry and exit points of the nucleosome
- DNA gyre
double stranded DNA wraps around the histone octamer in a left-handed manner leading to two adjacent DNA helices known as super-helical gyres
- Dyad
this axis passes through a single base-pair at the center of the nucleosomal DNA; the dyad straddles the H3:H3 interface and is denoted SHL 0
- H2A/H2B acidic patch
a region on the surface of the nucleosome enriched in acidic residues, composed of six residues from H2A (E56, E61, E64, D90, E91, E92) and two residues from H2B (E102, E110); frequently recognized by CAPs
- Histone H4 basic patch
a stretch of basic residues spanning Lys16-Arg23 in the N-terminus of H4
- Intrinsically disordered region (IDR) or protein (IDP)
a region of a protein, or a whole protein, which does not have the propensity to form stable secondary structure
- Nucleosome
the basic repeating unit of chromatin, composed of an octamer of histone proteins (two each of H2A, H2B, H3, and H4) wrapped by DNA; this is distinguished from the nucleosome core particle as it includes extra DNA than is required for the minimal wrap, termed linker DNA
- Nucleosome array
a series of nucleosomes all formed on a single piece of DNA
- Nucleosome core particle (NCP)
the histone octamer (two each of histones H2A, H2B, H3 and H4) wrapped by the minimal 146 or 147 base pairs of DNA needed for a full wrap
- Post-translational modifications (PTMs)
small chemical groups (or even small proteins) placed on proteins after their synthesis; these are usually placed on the side-chain or the very N- or C-terminus of the amino acid
- Reader domain
a small protein domain that can selectively bind to a modification state of a histone; examples are PHD fingers which recognize histone H3 either unmodified or methylated at Lys4, or bromodomains which recognize various acetylated lysines on histones
- Super helical location (SHL)
a specific DNA helical turn within the nucleosome core particle; the major grooves facing the histone core are numbered +1 through +7 and −1 through −7 either direction starting from the dyad (which is denoted 0), and the minor grooves are numbered in half-steps
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Olins DE and Olins AL (2003) Chromatin history: our view from the bridge. Nature Reviews Molecular Cell Biology 4, 809–814 [DOI] [PubMed] [Google Scholar]
- 2.Grigoryev SA (2012) Nucleosome spacing and chromatin higher-order folding. Nucleus 3, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henikoff S (2016) Mechanisms of Nucleosome Dynamics In Vivo. Cold Spring Harbor perspectives in medicine 6, a026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zentner GE and Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications. Nature structural & molecular biology 20, 259–266 [DOI] [PubMed] [Google Scholar]
- 5.Cutter AR and Hayes JJ (2015) A brief review of nucleosome structure. Febs Lett 589, [dummy_incompletepara] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song F et al. (2014) Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 [DOI] [PubMed] [Google Scholar]
- 7.Schalch T et al. (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 8.Kale S et al. (2019) Molecular recognition of nucleosomes by binding partners. Curr Opin Struc Biol 56, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews AJ and Luger K (2011) Nucleosome Structure(s) and Stability: Variations on a Theme. Annual review of biophysics 40, 99–117 [DOI] [PubMed] [Google Scholar]
- 10.Zhou K et al. (2018) Nucleosome structure and dynamics are coming of age. Nature structural & molecular biology DOI: 10.1038/s41594-018-0166-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierz B and Poirier MG (2019) Biophysics of Chromatin Dynamics. Annual review of biophysics DOI: 10.1146/annurev-biophys-070317-032847 [DOI] [PubMed] [Google Scholar]
- 12.Andrews FH et al. (2016) Insights into newly discovered marks and readers of epigenetic information. Nature chemical biology 12, 662–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhm L and Crane-Robinson C (1984) Proteases as structural probes for chromatin: the domain structure of histones. Bioscience Rep 4, 365–86 [DOI] [PubMed] [Google Scholar]
- 14.Zheng C and Hayes JJ (2003) Structures and interactions of the core histone tail domains. Biopolymers 68, 539–546 [DOI] [PubMed] [Google Scholar]
- 15.Bradbury EM et al. (1972) Nuclear-Magnetic Resonance and Optical-Spectroscopic Studies of Conformation and Interactions in the Cleaved Halves of Histone F2B. Eur J Biochem 26, 482–489 [DOI] [PubMed] [Google Scholar]
- 16.Feng Y et al. (2011) Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J Biological Chem 286, 20323–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X et al. (2000) Acetylation Increases the α-Helical Content of the Histone Tails of the Nucleosome. Journal of Biological Chemistry [DOI] [PubMed] [Google Scholar]
- 18.Kämpf K et al. (2018) What Drives 15N Spin Relaxation in Disordered Proteins? Combined NMR/MD Study of the H4 Histone Tail. Biophys J 115, 2348–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penna GL et al. (2006) Modeling H3 histone N-terminal tail and linker DNA interactions. Biopolymers 83, 135–147 [DOI] [PubMed] [Google Scholar]
- 20.Liu H and Duan Y (2008) Effects of Posttranslational Modifications on the Structure and Dynamics of Histone H3 N-Terminal Peptide. Biophys J 94, 4579–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cary PD et al. (1982) Effect of acetylation on the binding of N-terminal peptides of histone H4 to DNA. European journal of biochemistry / FEBS 127, 137–143 [DOI] [PubMed] [Google Scholar]
- 22.Hong L et al. (1993) Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biological Chem 268, 305–14 [PubMed] [Google Scholar]
- 23.Potoyan DA and Papoian GA (2012) Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. P Natl Acad Sci Usa 109, 17857–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K et al. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 25.Hansen JC (2002) Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Bioph Biom 31, 361–392 [DOI] [PubMed] [Google Scholar]
- 26.Pepenella S et al. (2014) Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma 123, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banères JL et al. (1997) The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. Journal of molecular biology 273, 503–508 [DOI] [PubMed] [Google Scholar]
- 28.Stützer A et al. (2016) Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Molecular Cell 61, 247–259 [DOI] [PubMed] [Google Scholar]
- 29.Zhou B-R et al. (2012) Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. Journal of molecular biology 421, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison EA et al. (2018) The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 7, e31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikebe J et al. (2016) H3 Histone Tail Conformation within the Nucleosome and the Impact of K14 Acetylation Studied Using Enhanced Sampling Simulation. PLoS computational biology 12, e1004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daban J-R and Cantor CR (1982) Role of histone pairs H2A,H2B and H3,H4 in the self-assembly of nucleosome core particles. J Mol Biol 156, 771–789 [DOI] [PubMed] [Google Scholar]
- 33.Ausio J et al. (1984) Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. Journal of molecular biology 176, 77–104 [DOI] [PubMed] [Google Scholar]
- 34.Iwasaki W et al. (2013) Contribution of histone N-terminal tails to the structure and stability of nucleosomes. Febs Open Bio 3, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurse NP et al. (2013) Clipping of flexible tails of histones H3 and H4 affects the structure and dynamics of the nucleosome. Biophys J 104, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andresen K et al. (2013) Solution scattering and FRET studies on nucleosomes reveal DNA unwrapping effects of H3 and H4 tail removal. Plos One 8, e78587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottesfeld JM and Luger K (2001) Energetics and Affinity of the Histone Octamer for Defined DNA Sequences †. Biochemistry-us 40, 10927–10933 [DOI] [PubMed] [Google Scholar]
- 38.Widlund HR et al. (2000) DNA Sequence-Dependent Contributions of Core Histone Tails to Nucleosome Stability: Differential Effects of Acetylation and Proteolytic Tail Removal †. Biochemistry-us 39, 3835–3841 [DOI] [PubMed] [Google Scholar]
- 39.Lilley DMJ and Tatchell K (1977) Chromatin core particle unfolding induced by tryptic cleavage of histones. Nucleic Acids Res 4, 2039–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira H et al. (2007) Histone Tails and the H3 αN Helix Regulate Nucleosome Mobility and Stability▿. Mol Cell Biol 27, 4037–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voltz K et al. (2012) Unwrapping of Nucleosomal DNA Ends: A Multiscale Molecular Dynamics Study. Biophys J 102, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenzaki H and Takada S (2015) Partial Unwrapping and Histone Tail Dynamics in Nucleosome Revealed by Coarse-Grained Molecular Simulations. Plos Comput Biol 11, e1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z and Kono H (2016) Distinct Roles of Histone H3 and H2A Tails in Nucleosome Stability. Scientific reports 6, 31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saikusa K et al. (2018) Structural Diversity of Nucleosomes Characterized by Native Mass Spectrometry. Anal Chem 90, 8217–8226 [DOI] [PubMed] [Google Scholar]
- 45.Karch KR et al. (2018) Hydrogen-Deuterium Exchange Coupled to Top- and Middle-Down Mass Spectrometry Reveals Histone Tail Dynamics before and after Nucleosome Assembly. Structure 26, 1651–1663.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann K et al. (2020) Dynamics of the nucleosomal histone H3 N-terminal tail revealed by high precision single-molecule FRET. Nucleic Acids Res 48, 1551–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaytan AK et al. (2015) Coupling between Histone Conformations and DNA Geometry in Nucleosomes on a Microsecond Timescale: Atomistic Insights into Nucleosome Functions. Journal of molecular biology DOI: 10.1016/j.jmb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy KJ et al. (2017) HMGN1 and 2 remodel core and linker histone tail domains within chromatin. Nucleic acids research 45, 9917–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullahoo J et al. (2020) Dual protease type XIII/pepsin digestion offers superior resolution and overlap for the analysis of histone tails by HX-MS. Methods DOI: 10.1016/j.ymeth.2020.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorigo B et al. (2003) Chromatin Fiber Folding: Requirement for the Histone H4 N-terminal Tail. J Mol Biol 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 51.Shogren-Knaak M et al. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 52.Bilokapic S et al. (2018) Cryo-EM of nucleosome core particle interactions in trans. Scientific reports 8, 7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kan PY et al. (2008) The H4 Tail Domain Participates in Intra- and Internucleosome Interactions with Protein and DNA during Folding and Oligomerization of Nucleosome Arrays. Mol Cell Biol 29, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pepenella S et al. (2014) A Distinct Switch in Interactions of the Histone H4 Tail Domain Upon Salt-Dependent Folding of Nucleosome Arrays. J Biol Chem 289, [dummy_incompletepara] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee K-M and Hayes JJ (1997) The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. … of the National Academy of Sciences [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K-M and Hayes JJ (1998) Linker DNA and H1-Dependent Reorganization of Histone−DNA Interactions within the Nucleosome†. Biochemistry-us 37, 8622–8628 [DOI] [PubMed] [Google Scholar]
- 57.Erler J et al. (2014) The role of histone tails in the nucleosome: a computational study. Biophys J 107, 2911–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakraborty K et al. (2018) Molecular Mechanism for the Role of the H2A and H2B Histone Tails in Nucleosome Repositioning. J Phys Chem B 122, 11827–11840 [DOI] [PubMed] [Google Scholar]
- 59.Fu I et al. (2017) Nucleosome histone tail conformation and dynamics: impacts of lysine acetylation and a nearby minor groove benzo [a]pyrene-derived lesion. Biochemistry DOI: 10.1021/acs.biochem.6b01208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Musselman CA et al. (2012) Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X and Hayes JJ (2007) Site-specific Binding Affinities within the H2B Tail Domain Indicate Specific Effects of Lysine Acetylation. Journal of Biological Chemistry 282, 32867–32876 [DOI] [PubMed] [Google Scholar]
- 62.Gatchalian J et al. (2017) Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 8, 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marabelli C et al. (2019) A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex. Cell Reports 27, 387–399.e7 [DOI] [PubMed] [Google Scholar]
- 64.Weaver T et al. (2018) Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 23, 2614–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilotto S et al. (2015) Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proc National Acad Sci 112, 2752–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morrison EA et al. (2017) DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun 8, 16080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maldonado R et al. (2019) Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells. Mol Cell 73, [DOI] [PubMed] [Google Scholar]
- 68.Bintu L et al. (2012) Nucleosomal elements that control the topography of the barrier to transcription. Cell 151, 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Újvári A et al. (2008) Histone N-terminal Tails Interfere with Nucleosome Traversal by RNA Polymerase II. J Biol Chem 283, 32236–32243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chatterjee N et al. (2011) Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res 39, 8378–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogler C et al. (2010) Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding. Plos Genet 6, e1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwarz M et al. (2018) Single-molecule nucleosome remodeling by INO80 and effects of histone tails. FEBS Letters 592, 318–331 [DOI] [PubMed] [Google Scholar]
- 73.Udugama M et al. (2011) The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Molecular and cellular biology 31, 662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Musselman CA et al. (2013) Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat Commun 4, 2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tencer AH et al. (2017) Covalent Modifications of Histone H3K9 Promote Binding of CHD3. Cell Reports 21, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrison EA et al. (2020) Nucleosome composition regulates the histone H3 tail conformational ensemble and accessibility. Biorxiv DOI: 10.1101/2020.06.26.172072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luger K et al. (2012) New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nature Reviews Molecular Cell Biology 13, 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hansen JC et al. (1998) Structure and Function of the Core Histone N-Termini: More Than Meets the Eye †. Biochemistry-us 37, 17637–17641 [DOI] [PubMed] [Google Scholar]
- 79.Howell SC et al. (2013) Elucidating internucleosome interactions and the roles of histone tails. Biophys J 105, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X and Hayes JJ (2007) Acetylation Mimics within Individual Core Histone Tail Domains Indicate Distinct Roles in Regulating the Stability of Higher-Order Chromatin Structure. Mol Cell Biol 28, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng C and Hayes JJ (2003) Intra- and Inter-nucleosomal Protein-DNA Interactions of the Core Histone Tail Domains in a Model System. J Biol Chem 278, 24217–24224 [DOI] [PubMed] [Google Scholar]
- 82.Dorigo B et al. (2004) Nucleosome Arrays Reveal the Two-Start Organization of the Chromatin Fiber. Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 83.Kaczmarczyk A et al. (2017) Single-molecule force spectroscopy on histone H4 tail-cross-linked chromatin reveals fiber folding. J Biol Chem 292, 17506–17513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu X et al. (2008) The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol 15, 1122–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu Q et al. (2011) Differential Contributions of Histone H3 and H4 Residues to Heterochromatin Structure. Genetics 188, 291–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nurse NP and Yuan C (2015) Cis and trans internucleosomal interactions of H3 and H4 tails in tetranucleosomes. Biopolymers 103, 33–40 [DOI] [PubMed] [Google Scholar]
- 87.Gao M et al. (2013) Histone H3 and H4 N-Terminal Tails in Nucleosome Arrays at Cellular Concentrations Probed by Magic Angle Spinning NMR Spectroscopy. J Am Chem Soc 135, [dummy_incompletepara] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luque A et al. (2016) Correlation among DNA Linker Length, Linker Histone Concentration, and Histone Tails in Chromatin. Biophys J 110, 2309–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang L and Takada S (2016) Histone acetylation dependent energy landscapes in tri-nucleosome revealed by residue-resolved molecular simulations. Scientific reports 6, 34441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saurabh S et al. (2016) Atomistic Simulation of Stacked Nucleosome Core Particles: Tail Bridging, the H4 Tail, and Effect of Hydrophobic Forces. J Phys Chem B 120, 3048–3060 [DOI] [PubMed] [Google Scholar]
- 91.Izadi S et al. (2016) Implicit Solvent Model for Million-Atom Atomistic Simulations: Insights into the Organization of 30-nm Chromatin Fiber. J Chem Theory Comput 12, 5946–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibson BA et al. (2019) Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bascom GD and Schlick T (2018) Chromatin Fiber Folding Directed by Cooperative Histone Tail Acetylation and Linker Histone Binding. Biophys J 114, 2376–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Collepardo-Guevara R et al. Chromatin Unfolding by Epigenetic Modifications Explained by Dramatic Impairment of Internucleosome Interactions: A Multiscale Computational Study. J Am Chem Soc 137, 10205–10215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27, 527–533 [DOI] [PubMed] [Google Scholar]
- 96.Tompa P and Fuxreiter M (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci 33, 2–8 [DOI] [PubMed] [Google Scholar]
- 97.Sharma R et al. (2015) Fuzzy complexes: Specific binding without complete folding. Febs Lett 589, 2533–42 [DOI] [PubMed] [Google Scholar]
- 98.Fuxreiter M (2018) Fuzziness in Protein Interactions—A Historical Perspective. J Mol Biol 430, 2278–2287 [DOI] [PubMed] [Google Scholar]
- 99.Rhee HS et al. (2014) Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell 159, 1377–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Musselman CA and Kutateladze TG (2019) Strategies for Generating Modified Nucleosomes: Applications within Structural Biology Studies. ACS Chemical Biology DOI: 10.1021/acschembio.8b01049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patel DJ (2016) A Structural Perspective on Readout of Epigenetic Histone and DNA Methylation Marks. Csh Perspect Biol 8, a018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soshnev AA et al. (2016) Greater Than the Sum of Parts: Complexity of the Dynamic Epigenome. Molecular Cell 62, 681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun Z-W and Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–8 [DOI] [PubMed] [Google Scholar]
- 104.Dover J et al. (2002) Methylation of Histone H3 by COMPASS Requires Ubiquitination of Histone H2B by Rad6. J Biol Chem 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 105.Sims JK et al. (2006) A Trans-tail Histone Code Defined by Monomethylated H4 Lys-20 and H3 Lys-9 Demarcates Distinct Regions of Silent Chromatin. J Biol Chem 281, 12760–12766 [DOI] [PubMed] [Google Scholar]
- 106.Ng HH et al. (2002) Ubiquitination of Histone H2B by Rad6 Is Required for Efficient Dot1-mediated Methylation of Histone H3 Lysine 79. J Biol Chem 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 107.Wu L et al. (2011) The RING Finger Protein MSL2 in the MOF Complex Is an E3 Ubiquitin Ligase for H2B K34 and Is Involved in Crosstalk with H3 K4 and K79 Methylation. Mol Cell 43, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Worden EJ et al. (2019) Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell 176, 1490–1501.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zheng S et al. (2010) Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Molecular and cellular biology 30, 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Furukawa A et al. (2020) Acetylated histone H4 tail enhances histone H3 tail acetylation by altering their mutual dynamics in the nucleosome. P Natl Acad Sci Usa DOI: 10.1073/pnas.2010506117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bowerman S and Wereszczynski J (2016) Effects of MacroH2A and H2A.Z on Nucleosome Dynamics as Elucidated by Molecular Dynamics Simulations. Biophysical journal 110, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Biswas M et al. (2011) Role of Histone Tails in Structural Stability of the Nucleosome. Plos Comput Biol 7, e1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lehmann K et al. (2017) Effects of charge-modifying mutations in histone H2A α3-domain on nucleosome stability assessed by single-pair FRET and MD simulations. Sci Rep-uk 7, 13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Borgia A et al. (2018) Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Danilenko N et al. (2019) Histone chaperone exploits intrinsic disorder to switch acetylation specificity. Nat Commun 10, 3435. [DOI] [PMC free article] [PubMed] [Google Scholar]


