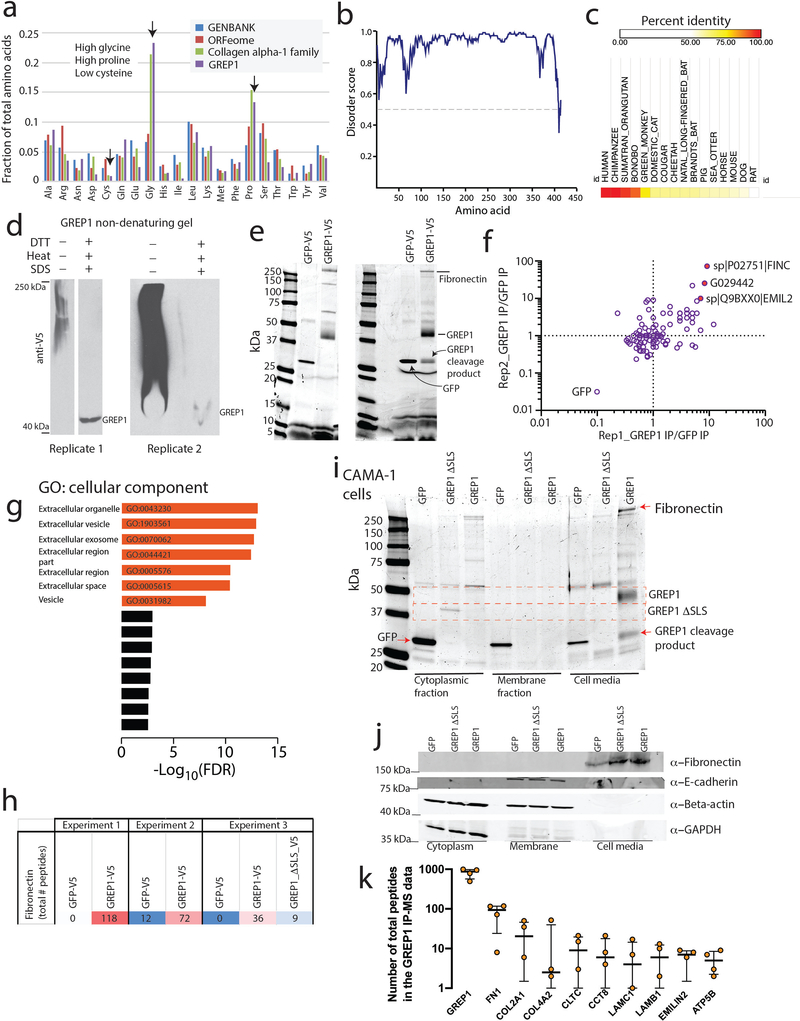
Extended Data Fig. 8. GREP1 is associated with the extracellular matrix.
a) Total fraction of amino acid usage in the ORFeome, GENBANK, GREP1, and the Collagen alpha-1 family. Sequence similarities between GREP1 and the collagen family are indicated. b) Predicted disorder score for the GREP1 amino acid sequence. c) Amino acid conservation for detected homologs of GREP1 in the indicated species. d) Non-denaturing native western blot of GREP1 in conditioned media from HEK293T cells expressing V5-tagged GREP1. e) Representative Commassie-stained gels for immunoprecipitation of GREP1 from the conditioned media of HEK293T cells. Two representative biological replicates are shown. f) Enrichment of extracellular matrix proteins in the IP-MS data for GREP1 compared to IP-MS data for GFP. g) Gene Ontology Cellular Component analysis of proteins >= 2 fold enriched in GREP1 immunoprecipitation compared to GFP immunoprecipitations. h) IP MS total peptide count for fibronectin shown for three separate experiments. i) Commassie stain of V5 immunoprecapitation of V5-tagged GFP, GREP1 del_SLS or GREP1 constructs expressed in CAMA-1 cells following fractionation of cell lysate into cytoplasmic, membrane and cell media components. Results were repeated in 2 independent experiments. j) Western blot of endogenous fibronectin, E-cadherin, beta-actin and GAPDH in cell lysate or cell culture media for CAMA-1 cells expressing GFP, GREP1 del_SLS or GREP1 constructs as in panel i. Results were repeated in two independent experiments. k) IP mass spectrometry data showing the total peptide count for GREP1 and other top-scoring proteins following IP of V5-tagged GREP1 in HEK293T, ZR-75–1, and CAMA-1 cells. N=4 independent IP MS experiments. Lines represent median +/− interquartile (25–75%) range.