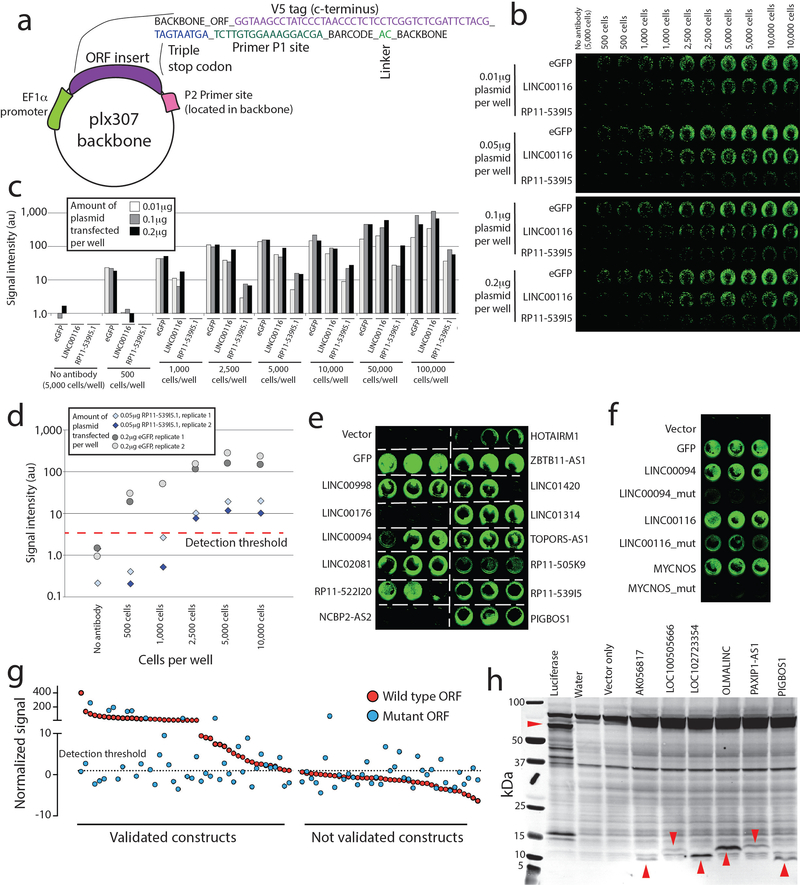
Extended Data Fig. 1. Generation and validation of a noncanonical ORF cDNA library.
a) Vector design and sequence details for the ORF library. The vector used is a modified version of the plx307 vector developed by the Genomic Perturbation Platform at the Broad Institute. b) Titration analyses of in cell western experiments. Three ORFs were chosen: eGFP (positive control), LINC00116 (high-expressing ORF), and RP11–539I5 (low expressing ORF). Increasing amounts of plasmid were transfected into increasing numbers of HEK293T cells as shown. c) Quantification the in cell western titration shown in b, demonstrating signal detection over noise and signal plateau. Signal was quantified using pixel density in the 800nM green color channel. d) Replicate experiments assessing signal-to-noise thresholds for a low-expressing ORF transfected into HEK293T cells with a low DNA plasmid concentration, as well as a high-expressing ORF (eGFP) transfected into HEK293T cells at a high DNA plasmid concentration. e) Example in cell western data in triplicate experiments for selected ORFs. f) Abrogation of protein translation via mutation of the ORF for selected examples. g) A systematic evaluation of in cell western signal for wild type and mutant ORFs for all pairs. ORFs are separated into those with signal above the baseline threshold, and those without reproducible signal. h) An immunoblot showing in vitro transcription/translation of selected tag-free ORFs using a wheat germ lysate system. Red arrows indicate the translated ORFs. Results were repeated in two independent experiments.