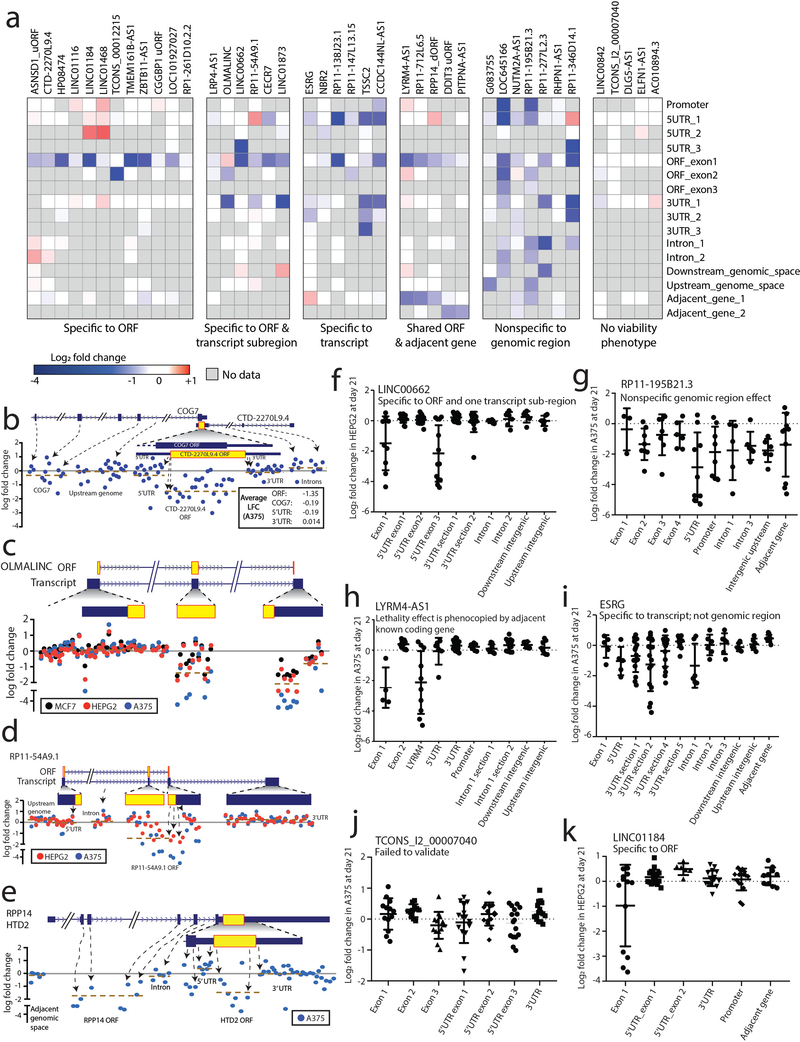
Extended Data Fig. 4. Tiling CRISPR assays to elucidate functional non-canonical ORFs.
a) A heatmap showing log fold change viability loss at Day +21 in the secondary CRISPR screen for the indicated non-canonical ORFs tested by multiple tiling sgRNA regions. b-e) Examples of non-canonical ORFs with a CRISPR tiling phenotype. b-e) Graphical representation of tiling CRISPR assays in which each dot represents an individual sgRNA. sgRNAs are mapped to their genomic loci and the genomic region of the tiling assay is shown. The location of the putative non-canonical ORF is shown in the gene annotation above. b) CTD-2270L9.4 c) OLMALINC d) RP11–54A9.1 e) RPP14 dORF / HTD2. f - k) Representative sgRNA log fold change data for the indicated transcripts. Each tiling experiment is classified as indicated. f) LINC00662 g) RP11–195B21.3 h) LYRM4-AS1 i) ESRG j) TCONS_I2_00007040 k) LINC01184.