Figure 5 |. TUT7 is a target of NAT1.
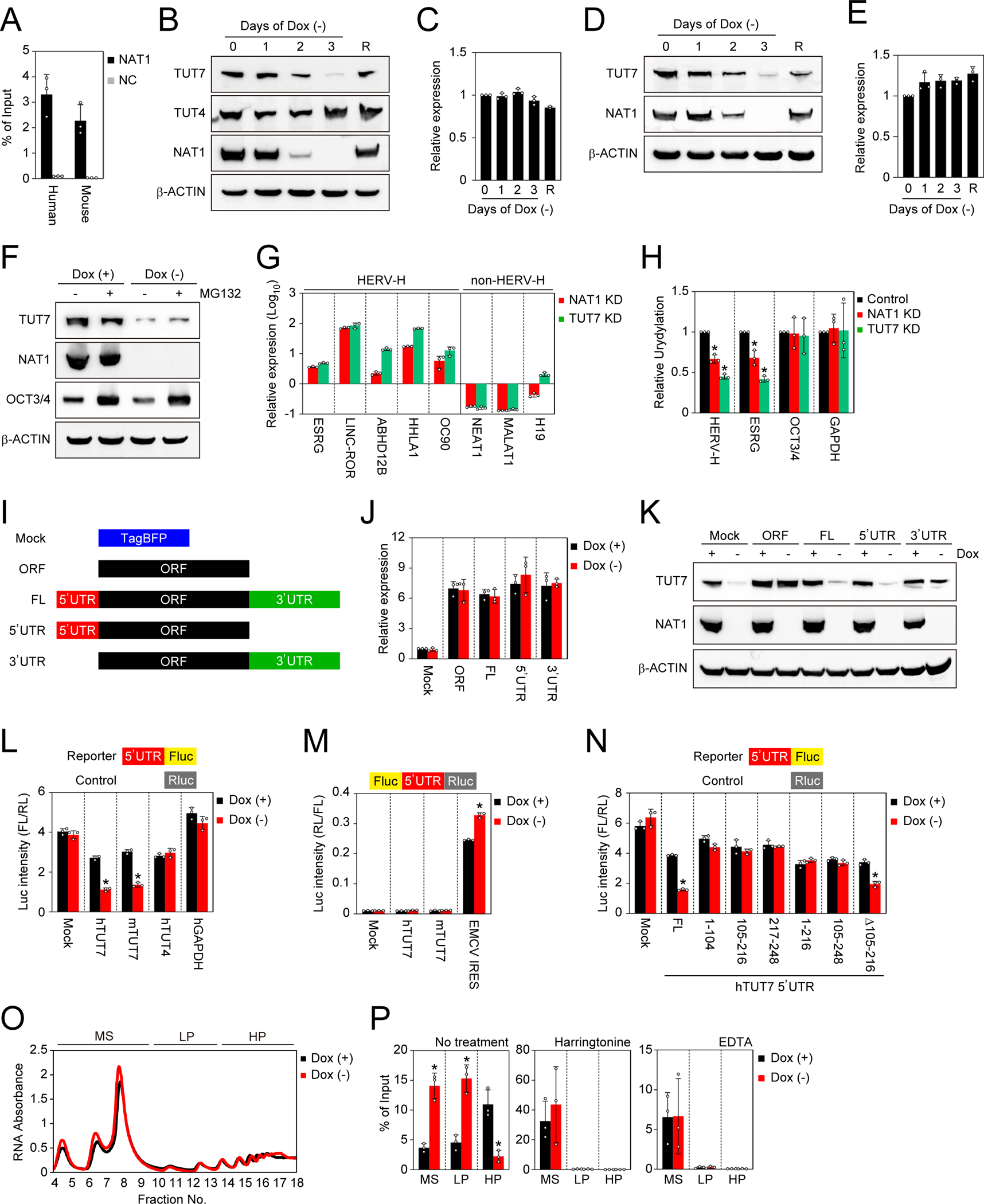
A. Pull down of TUT7 mRNA with NAT1 protein. TUT7 mRNAs in immunoprecipitants of 3xFLAG-tagged NAT1 (NAT1-FLAG) or non-tagged NAT1 (negative control, NC) human iPSCs and mouse EpiSCs purified using a FLAG antibody were quantified were by qRT-PCR. Values are normalized by input control. n=3. See also Tables S1 and S2.
B. Western blots of NAT1 cKO iPSCs in F/A condition on days 0–3 after Dox removal. On day 3 after Dox removal, we added Dox and collected the cell lysate after 1 day as rescued cells (R). See also Figure S4.
C. Relative expression of TUT7 in the cells shown in Fig. 5B analyzed by qRT-PCR. Values are normalized by GAPDH and compared with the sample on day 0. n=3.
D. Western blots of NAT1 cKO mouse EpiSCs on days 0–3 after Dox removal. On day 3 after Dox removal, we added Dox and collected the cell lysate after 1 day as rescued cells (R).
E. Relative expression of Tut7 in the cells shown in Fig. 5D analyzed by qRT-PCR. Values are normalized by Actb and compared with the sample on day 0. n=3.
F. Western blots of NAT1 cKO human iPSCs maintained in F/A condition with (+) or without (−) Dox and treated with (+) or without (−) MG132.
G. Relative expression of HERV-H and non-HERV-H non-coding RNAs in NAT1 KD or TUT7 KD human iPSCs in F/A condition analyzed by qRT-PCR. Values are normalized by GAPDH and compared with 1B4 human iPSCs. n=3. See also Figure S5.
H. Relative uridylation levels of pan HERV-Hs, ESRG, OCT3/4 and GAPDH RNAs in NAT1 KD or TUT7 KD human iPSCs in F/A condition analyzed by qRT-PCR Values are normalized by spike RNA and compared with the control (1B4). *P<0.05 vs. control by unpaired t-test. n=3. See also Figure S5.
I. Scheme of the TUT7 RNA dissection. See also Figure S6.
J. Relative expression of TUT7 in NAT1 cKO human iPSCs transfected with the plasmids shown in Fig. 5H maintained in F/A condition with (+) or without (−) Dox. Values are normalized by GAPDH and compared with Mock. n=3. See also Figure S6.
K. Western blots of the cells shown in Fig. 5J. See also Figure S6.
L. The effects of the UTRs on the activity of firefly luciferase (Fluc) in NAT1 cKO iPSCs in F/A condition with (+) or without (−) Dox. Values of Fluc are normalized with co-transfected Renilla luciferase (Rluc) activity. *P<0.05 vs. Dox (+) by unpaired t-test. n=3.
M. The effects of the UTRs on the activity of Rluc in NAT1 cKO human iPSCs in F/A condition with (+) or without (−) Dox. Values of Rluc are normalized with Fluc activity. *P<0.05 vs. Dox (+) by unpaired t-test. n=3. n. The truncation of TUT7 5’UTR. The effects of the series of truncated human TUT7 5’UTRs on the activity of Fluc in NAT1 cKO iPSCs in F/A condition with (+) or without (−) Dox were analyzed. Values of Fluc are normalized with co-transfected Rluc activity. *P<0.05 vs. Dox (+) by unpaired t-test. n=3.
O. Representative absorbance profiling of the total fractionated RNA of NAT1 cKO human iPSCs maintained with (+) or without (−) Dox. MS, monosomes (fraction 4–9), LP, light polysomes (fraction 10–13); HP, heavy polysomes (fraction 14–18).
P. The distribution of TUT7 mRNAs in fractionated MS, LP and HP of NAT1 cKO human iPSCs on days 0 (+) and 3 (−) of Dox removal treated with or without harringtonine or EDTA analyzed by qRT-PCR. Values are normalized by spike RNA and total input RNA, and compared with the sample on day 0. *P<0.05 vs. Dox (+) by unpaired t-test. n=3.
