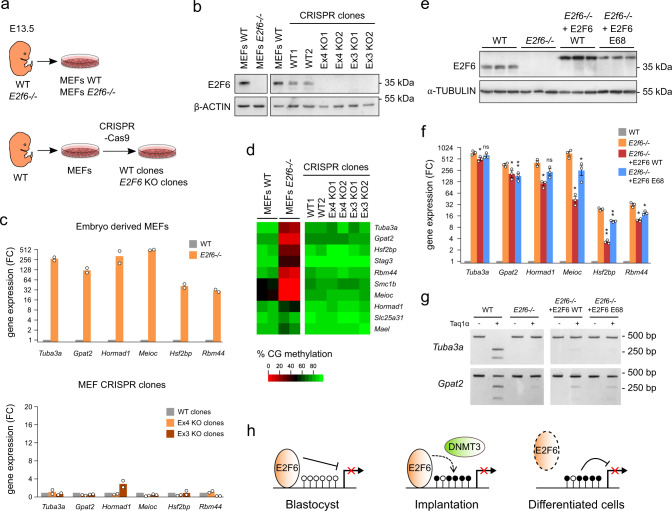
Fig. 6. E2F6 is dispensable for the maintenance of epigenetic silencing.
a Experimental design. We either derived MEFs from WT and E2f6−/− embryos or generated E2f6-KO clones and WT control clones by CRISPR-Cas9. b Western blotting of E2F6 in MEFs derived from WT and E2f6−/− embryos and MEFs CRISPR-Cas9 clones. β-ACTIN was used as a loading control. c RT-qPCR analysis of the expression of E2F6 target genes in embryo-derived WT and E2f6−/− MEFs (top), and MEF CRIPSR-Cas9 clones (bottom) (mean fold change compared to WT controls, n = 2 independent cell lines or clones per condition, expression normalized to Gusb, Rpl13a, and Mrpl32). d Heatmap representing the percentage of CpG methylation profiled by RRBS in the promoters of E2F6 target genes in embryo-derived WT and E2f6−/− MEFs (left), and MEF CRISPR-Cas9 clones (right). e Western blotting of E2F6 in WT and E2f6−/− MEFs, and E2f6−/− MEFs transduced with lentiviruses expressing E2F6 or E2F6 E68 fused to 3×-HA. α-TUBULIN was used as a loading control. f RT-qPCR analysis of the expression of E2F6 target genes in WT and E2f6−/− MEFs, and E2f6−/− MEFs rescued with E2F6. The values are represented as the fold change compared to WT (mean ± SEM, n = 3 independent experiments, expression normalized to Gusb, Rpl13a, and Mrpl32). *p < 0.05, **p < 0.01, ns: not significant (two-tailed t-test compared to E2f6−/− cells). g Methylation analysis by COBRA of the Tuba3a and Gpat2 promoters in WT and E2f6−/− MEFs, and E2f6−/− MEFs rescued with E2F6. h Model: E2F6 binds to and represses promoters of germline genes in preimplantation stages. At implantation, E2F6 recruits DNA methylation and initiates long-term epigenetic silencing of its target germline genes and subsequently becomes dispensable for the maintenance of the epigenetic repression in differentiated cells. Source data are provided as a Source Data file.