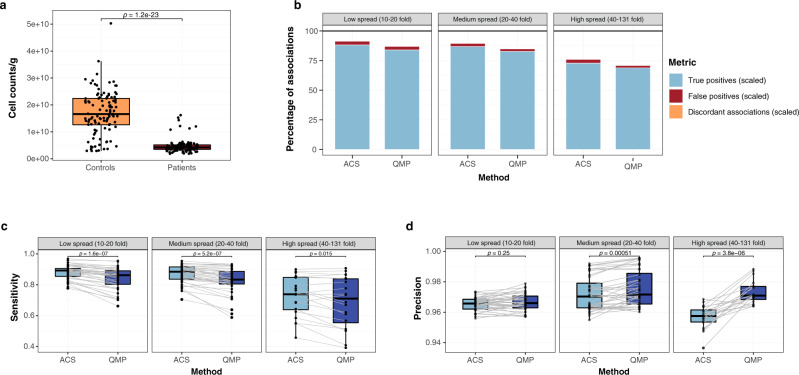
Fig. 5. Detection of disease-associated taxa by quantitative methods.
a Visualization of microbial loads distribution for samples assigned to the simulated patient and control groups in one of the dysbiosis matrices randomly selected (in which n = 200 samples, distributed as 93 healthy and 107 diseased; Wilcoxon rank-sum test, two-sided; p-adjusted for n = 10 matrices). The disease status variable is associated with total cell counts, but noise is present in the dataset. b Stacked barplots represent the percentage of true positive, false positive, and discordant taxon-disease associations recovered in ACS- and QMP-transformed sequence matrices at different sequencing depths, scaled to the number of taxon-disease associations present in the synthetic communities. c Sensitivity of both methods in detecting taxa-disease associations at different sequencing depths (Wilcoxon signed-rank test, two-sided). d Precision of both methods in detecting taxa-disease associations at different sequencing depths (Wilcoxon signed-rank test, two-sided). In b–d, simulated synthetic community matrices and their derived sequencing data were classified in three groups corresponding to the different spreads in microbial loads in the population (low spread, left panel: n = 40 sequencing matrices simulated from 4 synthetic communities; medium spread, mid panel: n = 40 sequencing matrices simulated from 4 synthetic communities; high spread, right panel: n = 20 sequencing matrices simulated from 2 synthetic communities). In a, c, d, the boxplots extend from the first to the third quartile of the distribution, with the line indicating the median. The whiskers cover from the quartiles to the last data point within 1.5x the interquartile range, with outliers depicted as individual points. ACS: absolute count scaling, QMP: quantitative microbiome profiling.