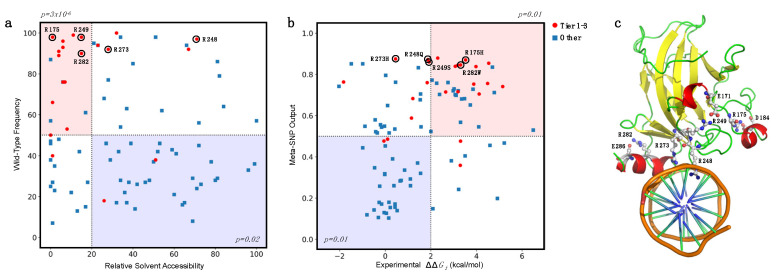
Figure 7.
Enrichment and depletion of putative cancer-driving variants (Tier 1–3) in different subgroups. (a) Enrichment of Tier 1–3 variants in mutated sites with Relative Solvent Accessibility ≤20% and frequency of the wild-type residue > 50% (light red). (b) Enrichment of Tier 1–3 variants in the subset of mutants with experimental ΔΔGf > 2.0 kcal/mol and Meta-SNP output >0.5 (light red). In both cases, the opposite regions (light blue) are depleted of Tier 1–3 variants. In (a,b), the hotspot mutants R175, R248, R249, R273, and R282 are highlighted with black circles. (c) Hotspot sites in the p53 structure in interaction with DNA (PDB:1TUP). Residues R248 and R273 interact with DNA. Residues R175, R249, and R282 are likely to stabilize the protein structure by forming salt bridge interactions with D162, E171, and E286, respectively.