Figure 6.
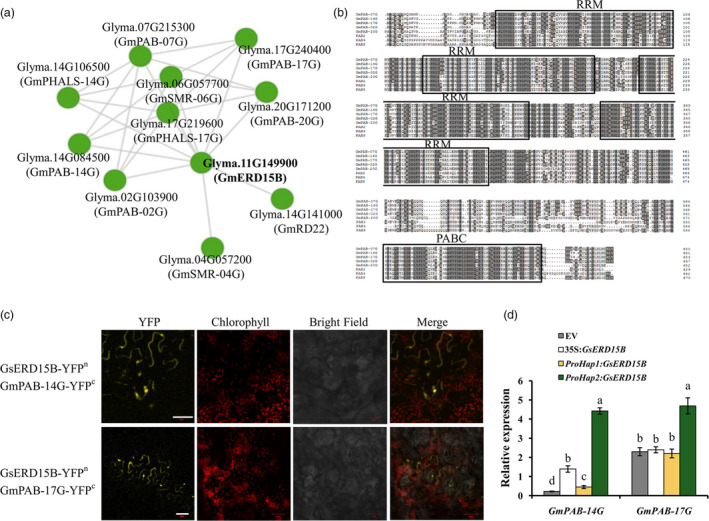
Prediction, sequence alignment, confirmation and relative expression of the GsERD15B‐interacting PAB proteins/genes. (a) The predicted protein–protein interaction network of GmERD15B using STRING database and Cytoscape software. Nodes represent proteins and edges represent interactions. (b) Sequence alignment of PAB proteins, including the five soybean PAB proteins that might interact with GmERD15B (GmPAB‐07G, GmPAB‐14G, GmPAB‐17G, GmPAB‐02G and GmPAB‐20G) and three known PAB proteins (PAB2, PAB4 and PAB8) from Arabidopsis, using full‐length amino acid sequence. The conserved RRM and PABC domains are indicated by boxes. (c) Bimolecular Fluorescence Complementation (BiFC) assay showing that GsERD15B (having the same sequence as GmERD15B) interacts with GmPAB‐14G, GmPAB‐17G in leaf cells of Nicotiana benthamiana. YFP, yellow fluorescent protein. Scale bar, 50 μm. (d) Relative expression levels of the two GmPAB genes at 3 h after 100 mM NaCl treatment in soybean (Tianlong1) hairy roots transformed by empty vector (EV, pBinGFP4), 35S:GsERD15B, ProHap1:GsERD15B, or ProHap2:GsERD15B, respectively. Root samples without salt stress (0 mM NaCl) for each genotype were used as the corresponding controls, and GmUKN1 was the reference gene. Data represent mean ± standard deviation of four biological replications (n ≥ 12). Same letters above bars for each gene indicate no significant difference between genotypes according to Duncan’s multiple range test at 0.05 level.
