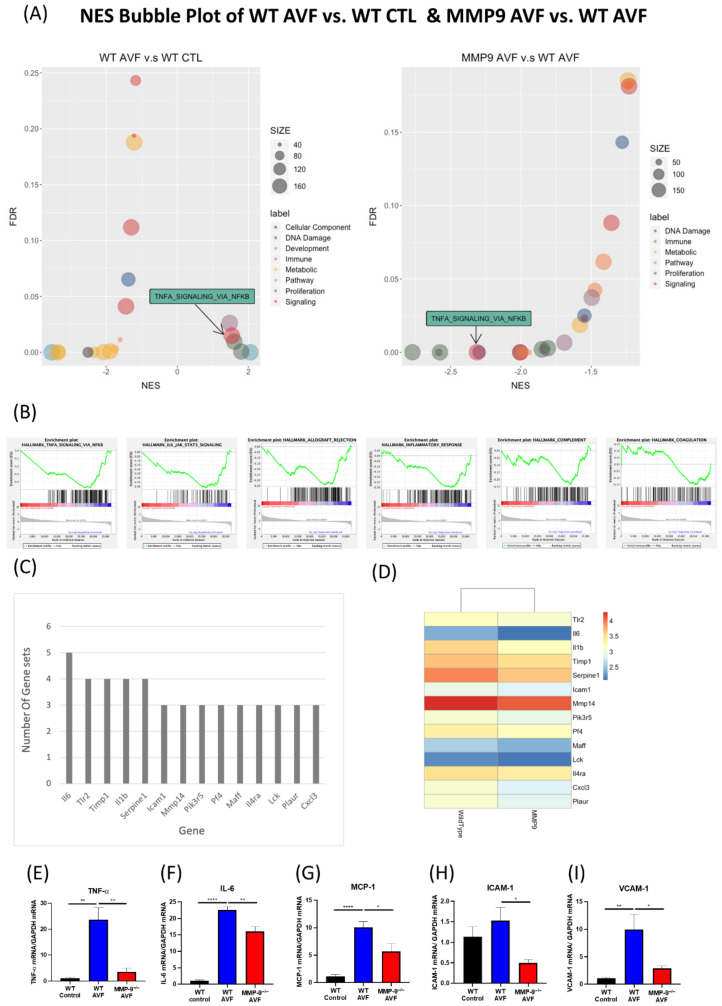
Figure 5.
Genome-wide mRNA expression difference of WT and MMP-9−/− mice AV fistulas. (A) Gene set enrichment analysis (GSEA) with Hallmark gene sets was performed to analyze the differentially expressed genes between WT and MMP-9−/− mice. Significantly enriched pathways in different categories are visualized using bubble plots of normalized enrichment score (NES) for different comparisons, including WT AVF vs. WT CTL and MMP9 AVF vs. WT AVF, and the size of each bubble is proportional to the number of core genes within the pathway. Only pathways with FDR < 0.25 were included. TNFA_SIGNALING_VIA_NFKB was positively enriched in WT AVF vs. WT CTL, but negatively enriched in MMP9 AVF vs. WT AVF. (B) GSEA enrichment plots of immune-related pathways in MMP-9−/− AVF vs. WT AVF. (C) Leading-edge analysis of six immune-related pathways. (D) The normalized expression value of these 14 key driver genes were expressed in heatmap, n = 3 in each group. Expression of inflammation-related genes usually associated with AV fistula stenosis such as (E) TNF-α, (F) IL-6, (G) MCP-1, (H) ICAM-1, and (I) VCAM-1, were examined by qPCR, n = 6–7 in each group. * p < 0.05, ** p < 0.01, **** p < 0.0001. Data presented as mean ± SEM. Data analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc analysis. ICAM-1, intercellular adhesion molecule 1; IL-6, interleukin 6; MCP-1, monocyte chemoattractant protein 1; NES, normalized enrichment score; NFKB, nuclear factor kappa-light-chain-enhancer of activated B cells; TNFA and TNF-α, tumor necrosis factor-α; VCAM-1, vascular cell adhesion molecule 1.