Abstract
Background
Microorganisms drive critical global biogeochemical cycles and dominate the biomass in Earth’s expansive cold biosphere. Determining the genomic traits that enable psychrophiles to grow in cold environments informs about their physiology and adaptive responses. However, defining important genomic traits of psychrophiles has proven difficult, with the ability to extrapolate genomic knowledge to environmental relevance proving even more difficult.
Results
Here we examined the bacterial genus Arthrobacter and, assisted by genome sequences of new Tibetan Plateau isolates, defined a new clade, Group C, that represents isolates from polar and alpine environments. Group C had a superior ability to grow at −1°C and possessed genome G+C content, amino acid composition, predicted protein stability, and functional capacities (e.g., sulfur metabolism and mycothiol biosynthesis) that distinguished it from non-polar or alpine Group A Arthrobacter. Interrogation of nearly 1000 metagenomes identified an over-representation of Group C in Canadian permafrost communities from a simulated spring-thaw experiment, indicative of niche adaptation, and an under-representation of Group A in all polar and alpine samples, indicative of a general response to environmental temperature.
Conclusion
The findings illustrate a capacity to define genomic markers of specific taxa that potentially have value for environmental monitoring of cold environments, including environmental change arising from anthropogenic impact. More broadly, the study illustrates the challenges involved in extrapolating from genomic and physiological data to an environmental setting.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-021-01084-z.
Keywords: Genomics, Metagenomics, Psychrophiles, Polar environment, Alpine environment, Microbial adaptation
Background
Many biotic and abiotic factors influence the ability of microorganisms to become indigenous members of environmental communities. Certain environmental factors can limit or prevent the growth of microorganisms, while enhancing, or being essential for others, resulting in ecological niches that support specific microbiome structures [1]. This phenomenon is well illustrated by a Winogradsky column where light and oxygen can be seen to exert major influences on the diversity and the dynamic of microorganisms throughout its length [2]. In more recent times, particularly through technological advancement (e.g., metagenomics), the understanding of microbial ecology and the contributions that microorganisms make to the natural world has grown considerably. Appreciation for microorganisms has accrued from discoveries of new biomes capable of supporting microbial colonization, such as the deep subsurface [3]; new examples of life hidden within microbial “dark matter” (e.g., Asgard archaea [4]); and the dynamic nature of microbial responses, particularly those that provide surprises, such as the major societal upheaval caused by the SARS-CoV-2 coronavirus. There is a growing realization that microorganisms constitute the life support system of the biosphere and must be properly accounted for when devising strategies to mitigate the impacts of human activity on the natural world [5]. In essence, we are living in a period in history when the need for society to learn about microbial responses to natural and anthropogenic influences is of unprecedented relevance [5–7].
Metagenomic methods have provided a level of insight into microbial communities [8, 9] that could possibly be equated to the advances made by Carl Woese and colleagues when using rRNA sequencing to discover Archaea as the third domain of life. Applied to the cold biosphere, Earth’s single largest biome, metagenomic analyses have catalogued diverse ways in which microbial life has evolved. As an example, Antarctic, marine-derived lake communities have been shown to have evolved independently over their relatively short history of 3000–5000 years, adapting not just to low temperature but also to a variety of important environmental factors specific to each lake system (reviewed in Ref. [10]). Metagenomic analyses have also begun to be used to uncover the ways in which communities in polar environments respond to changing environmental conditions; for example, the effects of the seasonal polar sunlight cycle on Antarctic marine and marine-derived lake communities [11, 12] and the roles that Arctic bacteria play in melting permafrost acting as a CO2 source and atmospheric CH4 sink [13, 14].
Arthrobacter (Actinobacteria; Micrococcales; Micrococcaceae) are a globally distributed genus of bacteria commonly found in soil, but also in a broad range of environments including water, human skin, and sewage [15–18]. Arthrobacter are reported to play important roles in global biogeochemical cycles and decontamination of polluted environments [17, 19]. Their responses to temperature, desiccation, ionizing radiation, oxygen radicals, and a range of chemicals have been described [20–22]. Their growth in the laboratory is characterized by nutritional versatility that translates to an ability to grow aerobically in media utilizing a wide range of carbon and nitrogen sources [16]. Some isolates closely related to the type species A. globiformis were obtained from a Lapland glacier region [23], and the identification of psychrophilic species has led to the characterization of a number of Arthrobacter enzymes for their biotechnological potential (e.g., Ref. [24]). Arthrobacter have been isolated from a range of low-temperature environments, including permafrost and glaciers [25–27].
Due to the large scale of the Earth’s cold biosphere and its relevance to global biogeochemical cycles, and the biotechnological potential of psychrophiles and their products, numerous studies have been performed to attempt to define the critical traits of psychrophiles (discussed in Ref. [10, 24, 28–34]). In the current study, we sequenced the genomes of Arthrobacter isolated from lakes, glaciers, and a wetland from the Tibetan Plateau (Additional file 1: Fig. S1) and utilized the existence of more than 100 Arthrobacter genomes to assess traits that may explain the presence of the genus in naturally cold environments. After identifying a clade characteristic of polar and alpine environments and determining that representatives had a superior ability to grow at low temperature, we used available metagenome data to assess the environmental relevance of the findings. What we learned illustrated the complexities involved in attempting to extrapolate from genomic and physiological data to an environmental setting. It also revealed possible avenues for utilizing Arthrobacter as biomarkers of environmental warming.
Results
Phylogenomics
To increase the number of Arthrobacter genomes from polar and alpine (PA) environments, a total of 16 isolates from seven lakes, two glaciers, and one wetland on the Tibetan Plateau (Additional file 1: Fig. S1 and Table S1) were sequenced (see the “Methods” section). After genome quality assessment and dereplication, 13 high-quality genome sequences for new Tibetan Plateau isolates were incorporated into the study (Additional file 2: Dataset S1). The phylogenomic relationships of a total of 210 non-redundant high-quality Micrococcaceae genomes (Additional file 2: Dataset S1) were analyzed by constructing maximum likelihood and Bayesian trees (Additional file 1: Fig. S2). The two trees were congruent and most tree-nodes (194/208) were supported by high bootstrap values (>70%) (Additional file 1: Fig. S2). The Arthrobacter lineage formed a cluster with 106 representatives that was clearly separated from other Micrococcaceae genera (Additional file 1: Fig. S2). The Arthrobacter genomes represented PA isolates (total 31, including the 13 new genomes), with the remainder from a broad range of non-polar or alpine (NPA) environments (Additional file 2: Dataset S1).
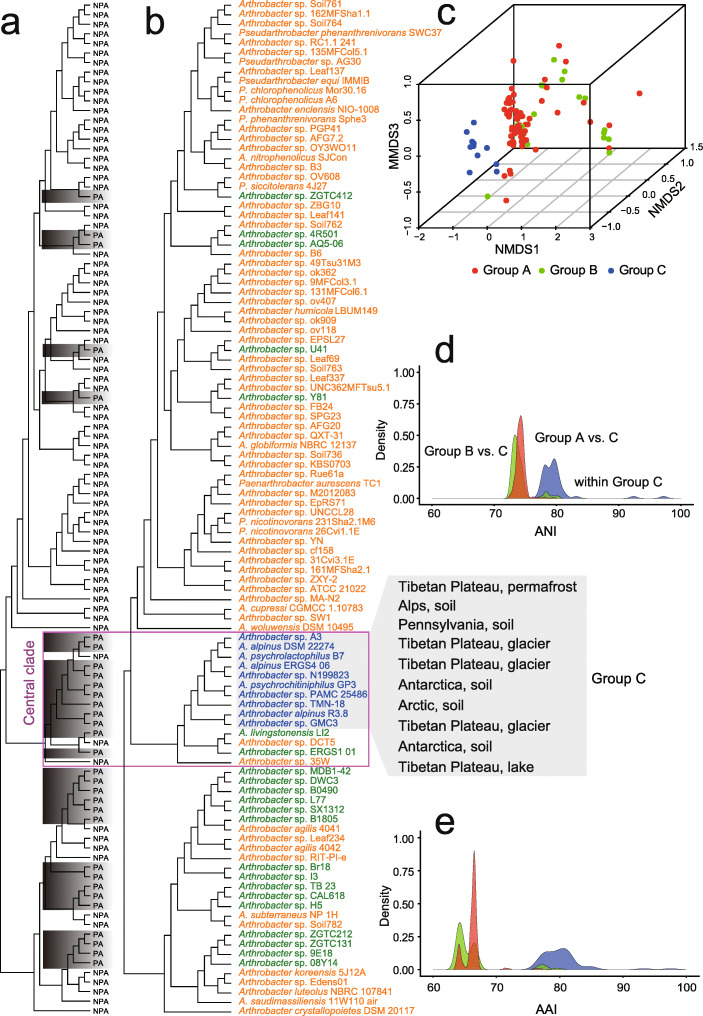
The Arthrobacter lineage separated into three clades that branched from the root of the Arthrobacter tree (Fig. 1). The 31 PA Arthrobacter were distributed across the tree, although 11 PA Arthrobacter formed a cluster with three NPA Arthrobacter in the central clade (Fig. 1 a). Within the central clade, 10 Arthrobacter grouped together (Fig. 1 b, blue font) with an F measure of 0.95, defining them as an operationally monophyletic lineage [35]. This was supported by three-dimensional nonmetric multidimensional scaling analysis of amino acid composition (Fig. 1 c) and both average nucleotide identity (ANI) and average amino acid identity (AAI) distributions (Fig. 1 d, e). The grouping of 10 Arthrobacter (Fig. 1 b, blue font) consisted of nine PA Arthrobacter, plus A. psychrolactophilus B7 which was isolated from Pennsylvania soil following snow melt; the isolate was obtained as a source of cold-active enzymes and was capable of growth at 0 °C [36]. The monophyletic lineage of 10 Arthrobacter was defined as Group C (Fig. 1 b, blue font), with all other PA Arthrobacter as Group B (Fig. 1 b, olive green font), and all Arthrobacter from NPA environments as Group A (Fig. 1 b, orange font).
Fig. 1.
Arthrobacter phylogeny and genome compositional profiling. a Maximum likelihood Arthrobacter phylogenomic tree. The Arthrobacter portion of maximum likelihood Micrococcaceae phylogenomic tree (Additional file 1: Fig. S2a) is reproduced with each leaf marked as polar and alpine (PA, gray highlight) or non-polar and alpine (NPA). The tree has three major clades with the central clade highlighted (purple box). b As for a except Arthrobacter names denoted and font color used to depict Group A (orange font; NPA environments), Group B (olive green font; PA environments clustering with sequences from NPA environments), and Group C (blue font; PA environments that formed an operationally monophyletic lineage with an F measure of 0.95). The specific types of cold environments from where Group C Arthrobacter were isolated are shown to the right of the tree. c Three-dimensional nonmetric multidimensional scaling (NMDS) plot of genome-wide amino acid composition. d Distribution of pairwise average nucleotide identity (ANI). e Distribution of pairwise average amino acid identity (AAI)
Low-temperature growth capacity of Group C Arthrobacter
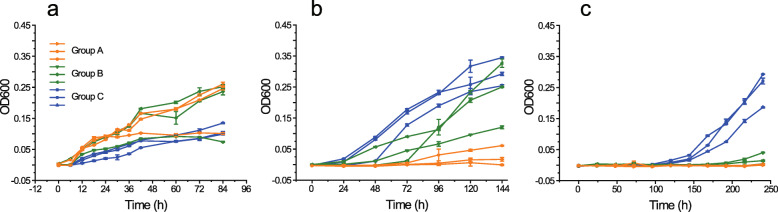
To evaluate the growth temperature response of Group A, B, and C, three Arthrobacter from each group were grown at 25, 5, and −1 °C and growth monitored (OD600) over time (Fig. 2). The three Group C Arthrobacter exhibited a markedly enhanced rate of growth at −1 °C (Fig. 2 c) particularly compared to Group A Arthrobacter and had a reduced rate of growth at 25 °C compared to some of the Group A and B Arthrobacter (Fig. 2 a, b).
Fig. 2.
Growth temperature profiles of Group A, B, and C Arthrobacter. OD600 growth curves for representative Arthrobacter of Group A (orange symbols and line; A. luteolus, A. globiformis, and A. subterraneus), Group B (olive green symbols and line; Arthrobacter sp. 4R501, Arthrobacter sp. 9E14, and Arthrobacter sp. 08Y14), and Group C (blue symbols and line; A. alpinus, Arthrobacter sp. A3, and Arthrobacter sp. N199823) at a 25 °C, b 5 °C, and c −1 °C
Genomic characteristics
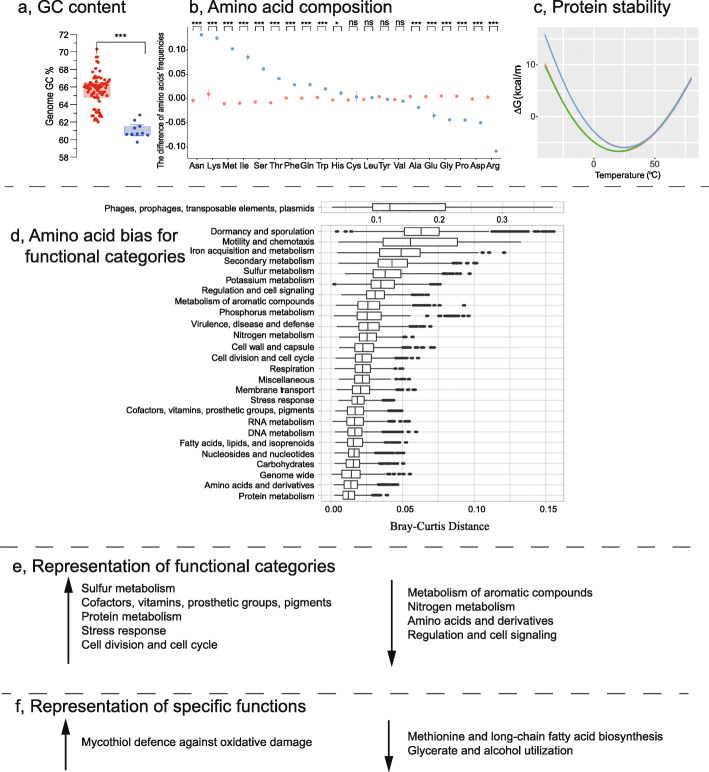
The size of the 106 Arthrobacter genomes ranged from 3.24 to 5.89 Mbp (Additional file 2: Dataset S1). Between Group A and C, no significant differences occurred in genome size, 16S rRNA and tRNA gene copy number, or coding density (Additional file 1: Fig. S3). However, a significant difference was observed in amino acid composition and G+C content (Fig. 3, Additional file 1: Fig. S3 and Additional file 2: Dataset S1). In Group C, the content of N, K, M, I, S, T, F, Q, W, and H was significantly higher, while A, E, G, P, D, and R was significantly lower (one-way ANOVA, p < 0.05, Additional file 1: Fig. S4). The correlation between amino acid composition and G+C content was significantly negative for the amino acids N, I, M, S, F, K, and Q (R2 ranged from 0.46 to 0.81; p < 0.01) and significantly positive for W, G, D, P, R, and A (R2 ranged from 0.39 to 0.77; p < 0.01) (Additional file 1: Fig. S5).
Fig. 3.
Overview of genomic characteristics of Group C Arthrobacter. a Box plot of G+C content. Group A (red); Group C (blue); boxes represent the interquartile range with horizontal lines showing maximum and minimum values, excluding outliers. Group C had significantly lower G+C content. b Scatter plot of amino acid composition. Group A (light red circles); Group C (blue circles); ***p < 0.005; *p = 0.05–0.01; ns, not significant. The composition of numerous amino acids varied significantly between Group C and Group A Arthrobacter. c Protein stability predictions calculated using SCooP. Group A (red line); Group C (blue line). The curve is for coenzyme A biosynthesis bifunctional protein, CoaC, and is representative of one of the 32 Group C proteins from a total of 86 which had reduced predicted stability (Additional file 3: Dataset S2 and Additional file 1: Fig. S6) d Box plot of amino acid bias for functional categories. Boxes represent the interquartile range of the Bray-Curtis distances; lines extending from boxes show the maximum and minimum Bray-Curtis distances; dots beyond the lines represent outliers. Biases in amino acid composition (b) were reflected in specific functional categories. e Representation of functional categories. Specific functional categories were over- or under-represented in Group C; arrows indicate relative increases (up arrow) or decreases (down arrow) in functional categories in Group C. f Representation of specific functions. Specific functional processes defined by genes or pathways were characteristic of Group C (up arrow) or had a restricted capacity in Group C (down arrow) compared to Group A (also see Fig. 4)
To evaluate the potential structural relevance of the amino acid compositional differences, temperature-dependent protein stability predictions were made using SCooP, which predicts stability assuming proteins are monomeric and follow a two-state folding transition [37]. From 180 proteins targeted for evaluation (mostly single-copy genes; see the “Methods” section), 86 produced robust stability curves (Fig. 3, Additional file 1: Fig. S6 and Additional file 3: Dataset S2). Stability was calculated at −1 °C to match the growth temperature at which Group C showed a marked difference in growth ability (Fig. 2). The 86 Group C proteins had significantly higher ΔG values (Group A, −4.27; Group B, −4.29; Group C, −3.60; p < 0.01), with 32 proteins being responsible for the reduced predicted stability (Additional file 1: Fig. S6 and Additional file 3: Dataset S2). These 32 proteins contained a particularly high representation of the amino acids that were most over-represented in Group C (i.e., N, K, and R; Additional file 1: Fig. S4). The 32 proteins represented 12 functional categories, primarily metabolism (28 proteins; 9 categories), with four involved in respiration, stress response, or cell division and cell cycle (Additional file 3: Dataset S2). The marked amino acid compositional differences, broad representation of functional categories, and high proportion of proteins with predicted decreases in stability (~ 1/3rd of those tested) demonstrate that Group C Arthrobacter possess broad genomic differences to Group A Arthrobacter. If the decreases in predicted protein stability translate to an increased capacity to perform catalysis at low temperature, this may contribute to the higher growth rates of Group C at −1°C (Fig. 2).
To further explore the influence of amino acid composition on functional potential, Bray-Curtis distances of genome-wide amino acid composition were evaluated for proteins representing 26 functional categories (Fig. 3). The greatest distance was for the category “phages, prophages, transposable elements, plasmids,” consistent with previous studies associating transposable elements with cold-adapted microorganisms [38–40]. The functional potential of Group C was also compared to Group A using enrichment analysis [41] performed on proteins representing the 26 functional categories. Group C was over-represented in sulfur metabolism; cofactors, vitamins, prosthetic groups, pigments; protein metabolism; stress response; cell division; and cell cycle (Fig. 3, Additional file 1: Fig. S7 and Additional file 4: Dataset S3), whereas Group A was over-represented in the metabolism of aromatic compounds; nitrogen metabolism; amino acids and derivatives; regulation; and cell signaling (Additional file 1: Fig. S7 and Additional file 4: Dataset S3). The category “sulfur metabolism” also exhibited signatures of amino acid bias (Bray-Curtis distance; Fig. 3), suggesting selection for this functional capacity occurred at the levels of both gene complement and amino acid composition.
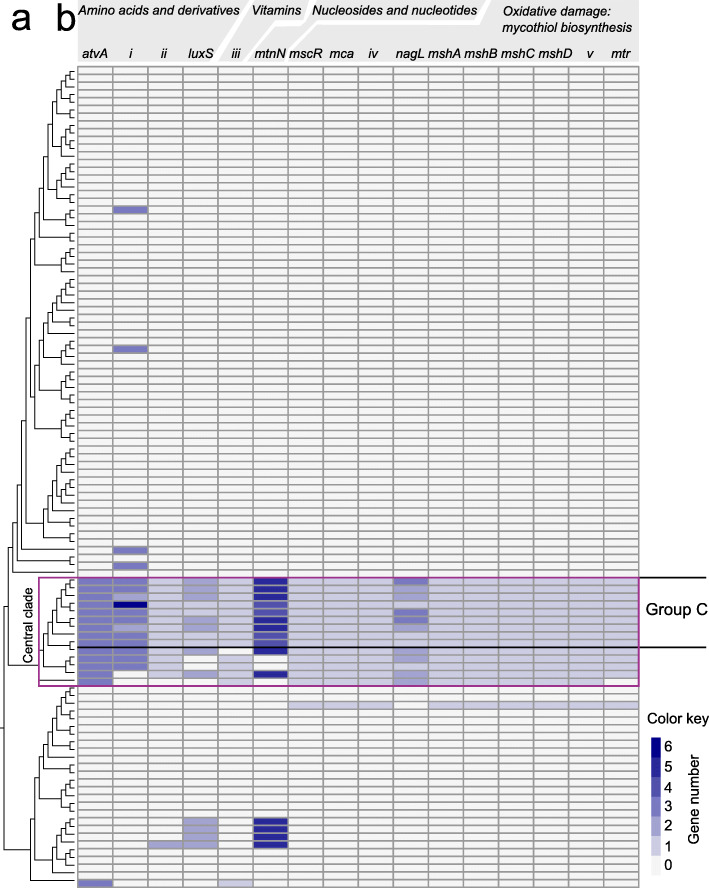
Functional assessments were extended to identify specific genes unique to Group C. A number of genes involved in the synthesis of amino acids, vitamins, and nucleosides were present in all Group C genomes (Fig. 4). The specific genes also tended to be present in the other four, non-Group C members (Group A and Group B) of the central clade (Fig. 4), but had low representation in other Arthrobacter genomes (Fig. 4). The most marked feature was a complete mycothiol (MSH) biosynthesis pathway that was present in all Group C genomes (Fig. 4); MSH is a redox-active thiol, functionally analogous to glutathione (which is typically absent from Actinobacteria), that maintains intracellular redox balance and can therefore protect against oxidative damage [42]. Furthermore, MSH potentially serves as a stable reservoir of carbon and sulfur in bacteria [43]. The ability to respond effectively to oxidative damage may be an important trait of microorganisms from cold environments, particularly for facilitating growth at low-temperature limits [44–48]. The MSH pathway was also present in the other four, non-Group C members of the central clade, plus one other Group B member (Fig. 4). Therefore, the MSH pathway plus the individual genes involved in the synthesis of amino acids, vitamins, and nucleosides are characteristic of Group C Arthrobacter, but are not unique to this group. If MSH or the other individual genes fulfill roles in facilitating growth at low temperature, the genes may be under stronger positive selection in Group C, while also being retained within Group A and Group B populations (pan genome), but at a significantly lower level.
Fig. 4.
Arthrobacter genes typifying the functional potential of Group C. a Maximum likelihood Arthrobacter phylogenomic tree as for Fig. 1. b Heat map of the representation of specific genes in Arthrobacter genomes, highlighting those present in Group C and the central clade. i, branched-chain acyl-CoA dehydrogenase; ii, enoyl-CoA hydratase; iii, biotin repressor; iv, hydrolase in cluster with formaldehyde/S-nitrosomycothiol reductase; v, mycothiol-dependent formaldehyde dehydrogenase
A total of 48 Group C gene families had significantly higher, and 66 had significantly lower average gene copy number compared to Group A, including four which were absent in Group C genomes (Additional file 5: Dataset S4, p < 0.05). The absence of two specific genes is noteworthy: adenosylhomocysteinase, which hydrolyzes S-adenosyl-l-homocysteine (a product of methyl transfer reactions that involve S-adenosyl-l-methionine) to homocysteine and adenosine [49], and formate–tetrahydrofolate (THF) ligase, which catalyzes the initial recruitment of single carbon units for THF-mediated one-carbon metabolism [50]. The absence of both genes would be expected to disrupt the synthesis of methionine from homocysteine, and instead favor the alternative pathway of synthesizing methionine from cysteine; the latter pathway may be connected to MSH metabolism, in that accumulation of cysteine (a precursor of MSH synthesis) is toxic to cells [43], so surplus cysteine not required for MSH synthesis, or resulting from MSH degradation, could be directed to methionine synthesis (Additional file 5: Dataset S4).
Some of the gene families had particularly high copy numbers per genome (~30 in Group A) with large reductions (~6) in Group C (Additional file 5: Dataset S4); this trend was observed for 3-oxoacyl-[acyl-carrier protein] reductase (FabG), glycerate kinase (GlxK), and alcohol dehydrogenase (Adh). For FabG, this likely reflects a reduced capacity of Group C to catalyze the formation of long-chain fatty acids (Additional file 5: Dataset S4). GlxK is an important catabolic enzyme, in that diverse substrates are degraded to glycerate, and GlxK links these degradation pathways to central carbon metabolism [43]. Decreased copy numbers of Adh likely indicates decreased capacities to utilize alcohols. Thus, decreases in GlxK and Adh might reflect decreased substrate preferences by these Arthrobacter. It was noteworthy that the copy number of cold shock protein (csp) genes was lower in Group C. While csp genes are sometimes equated with an ability to grow in the cold or survive cold shock, these nucleic acid binding proteins can perform diverse roles in cellular function (reviewed in Ref. [34]); the findings here reinforce the notion that csp and other “stress” genes are not good molecular markers for identifying psychrophiles [5, 33].
Ecology of Group C Arthrobacter
We hypothesized that if the laboratory-generated growth data (Fig. 2) and genomic traits (Fig. 3) translated to competitiveness in cold environments, Group C Arthrobacter would be over-represented in metagenome data from PA vs NPA environments. The relative abundance of Arthrobacter in environmental samples (publicly available metagenome data) tends to be low, with no metagenome-assembled genomes (MAGs) present in the ~8000 MAGs that were constructed from ~1500 metagenomes [51], and a total of 12 (> 90% completeness) present in 76,831 Integrated Microbial Genomes (IMG) MAGs (December 2019). To facilitate metagenome analyses, group-specific genes (Additional file 6: Dataset S5) were examined in 639 metagenomes representing PA, temperate, and tropical environments (Additional file 7: Dataset S6), with representation shown relative to the Arthrobacter pan genome (Fig. 5a) (see the “Methods” section for a description of the analytical approach).
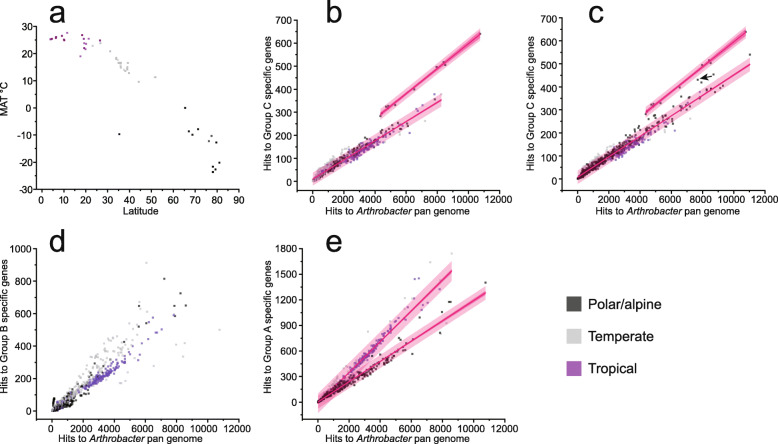
Fig. 5.
Metagenome analysis of Group C Arthrobacter. a Depiction of the mean annual temperature (MAT) of surface air at a height of 2 m (European Centre for Medium-Range Weather Forecasts) relative to latitude. The 639 metagenomes are divided into thermal categories: PA (black squares, 196 metagenome), temperate (gray squares, 243 metagenomes), and tropical (purple squares, 200 metagenomes). b Linear regression showing the correlation of the abundance of Group C-specific genes within each of the 639 metagenomes (see panel a) relative to the abundance of Group C-specific genes within the Arthrobacter pan genome. The 95% prediction interval (dark pink band) and 95% confidence interval (light pink band) are shown for each regression line (panels b, c, and e). The upper cluster contains 11 Axel Heiberg Island permafrost metagenomes. c As for panel b, except with the addition of 334 permafrost metagenomes (total 973 metagenomes). The Stordalen Mire (Abisko, Sweden) metagenome is shown by an arrow. d As for panel b, except showing Group B-specific genes. e As for panel b, except showing Group A-specific genes present in PA genomes (lower line) and NPA genomes (upper line). The regression line for the 11 Axel Heiberg Island permafrost metagenomes is not shown
Group C-specific genes were more highly represented in 11 permafrost metagenomes (Fig. 5b). All of the 11 metagenomes came from a single site: Axel Heiberg Island, Nunavut, Canada [52]. The Axel Heiberg Island study reported 76 metagenomes derived from 1-m cores that were used during a controlled thawing experiment [52]. Most Group C-specific genes were enriched in the 65-cm depth active-layer (7 metagenomes), with one from the 35-cm active-layer and three from the 80-cm permafrost-layer (Additional file 1: Table S2). A total of 94% of the variability that exists in hits to Group C for the 639 metagenomes (Fig 5b) was traced to pre-existing variability in hits to the Arthrobacter pan genome, and when this covariance was removed by ANCOVA analysis, a statistically significant difference in the y-intercepts for the regression lines (p < 0.0001) remained; this confirms the over-representation of Group C-specific genes in the 11 metagenomes compared to the remaining 628 metagenomes.
To assess whether Group C Arthrobacter were generally enriched in permafrost regions, all other publicly available permafrost metagenomes (334 metagenomes) were analyzed (Fig. 5c). A number appeared somewhat enriched in Group C-specific genes (e.g., a metagenome from Stordalen Mire near Abisko, Sweden; marked by an arrow in Fig. 5c), but as the slopes of the two regression lines were not parallel, it was not valid to compare the y-intercepts, and hence, the significance of the difference between them could not be evaluated [53].
For Group B-specific genes, no obvious trends separated the PA from NPA metagenomes (Fig. 5d). However, the distribution of Group A-specific genes clustered according to climate classification, with PA metagenomes showing lower Group A content (y = 0.1202x + 3.4193, R2 = 0.96801) compared to temperate and tropical metagenomes (y = 0.1804x − 15.015, R2 = 0.96989) (Fig. 5e). The 11 Axel Heiberg Island metagenomes had a statistically significant under-representation of Group A-specific genes compared to all other metagenomes (ANCOVA, p < 0.0001). This pattern indicates there is selection against Group A Arthrobacter in PA environments and/or selection for Group A in NPA environments.
To define variables that may explain the niche adaptation of Group C in the Axel Heiberg Island permafrost, available abiotic and biotic data were used from the permafrost study [13, 14]. A range of physicochemical data were available for each of the four depths (5, 35, 65, and 80 cm), but as the timing of sampling for physicochemical data (0, 4, 6, 8, 11, and 12 weeks) did not align with the timing of sampling for the metagenomes (0, 0.25, 6, 12, and 18 months), the physicochemical data were ultimately not useful for interpreting Group C distribution. Depth, treatment group, and sample core did not explain the variation in species composition across the sites, and although incubation time had some explanatory power for the distribution of the entire permafrost study dataset (data not shown), the metagenomes enriched in Group C Arthrobacter were widely distributed and did not cluster together, suggesting the importance of specific microniches in the enrichment of these species. Assessment of the functional potential of the microbial communities in each of the 76 metagenomes using the presence/absence of KO groups also did not identify any significant functional differences (PERMANOVA, p > 0.05; data not shown).
In contrast, strong taxonomic associations were identified with many members of the microbial community. Analyses were performed to assess taxa that correlated with Group A and Group C, just Group A, and just Group C (Additional file 8: Dataset S7). A total of 107 operational taxonomic units (OTUs) positively correlated with Group C Arthrobacter, and 63 negatively correlated (Additional file 8: Dataset S7). Of the 107 OTUs positively correlated to Group C, 72 were also positively correlated to Group A above the threshold of 0.5 (the remainder were positively correlated with values 0.359–0.499), and no OTUs were positively correlated to Group A that did not also correlate to Group C. The positively correlating OTUs were dominated by both spore- and non-spore-forming members of Actinobacteria and Firmicutes, as well as members of Proteobacteria; the majority of these OTUs were isolated from soil. Negatively correlating bacterial OTUs mainly belonged to marine or lacustrine members of Bacteroidetes, Cyanobacteria, and Proteobacteria, as well as certain eukaryotes (fungi, plants, marine annelid worm).
Discussion
Numerous studies have been performed to define the critical traits of a psychrophile, including those that have compared genomes that represent a broad range of species and thermal environments (discussed in Ref. [33, 34, 54]). The current study explored genomic characteristics of a lineage with less than 3.5% difference in 16S rRNA gene identity. The analyses revealed that the Arthrobacter lineage contains a clade with members (Group C) possessing a clear capacity to grow faster than their relatives (Group A and B) under laboratory growth conditions at −1 °C (Fig. 2). A number of genomic characteristics that potentially explain the physiological capacity of Group C were identified. (1) Group C possess an amino acid composition that is predicted to reduce the stability of a large proportion of proteins thereby enhancing enzyme activity at low temperature [53]. (2) Group C genomes are enriched in sulfur metabolism genes, and sulfur is required for the cysteine component of mycothiol. The synthesis of mycothiol may potentially protect Group C Arthrobacter against oxidative damage that may otherwise accumulate as cell division decreases towards the lower temperature limit of growth [44]. (3) Group C exhibits a relatively high proportion of mobile elements, which is a trait shared with some other cold-adapted microorganisms [38–40]. Collectively, the physiological and genomic traits appear compelling for denoting Group C, a cold-adapted clade of Arthrobacter.
However, from assessing available metagenome data, we infer that these traits do not translate to a generally enhanced ability to compete in low-temperature environments. Other than the 11 specific Axel Heiberg Island permafrost metagenomes, Group C Arthrobacter were not highly represented in the other metagenomes from cold environments, including 144 from Arctic peat soil, 22 associated with glaciers, 42 from polar deserts, and importantly, 365 from other permafrost environments. Even at the Axel Heiberg Island site, Group C-specific genes were not highly abundant at 5- and 20-cm depths. Instead, the pattern of abundance of Group C appears to derive not just from low temperature, but from niche-specific conditions.
Attempting to identify specific niche conditions is not trivial. For the Axel Heiberg Island study, the permafrost microbial community was reported to be dominated by Actinobacteria and Proteobacteria, with significant increases at depth for Firmicutes and Actinobacteria and significant decreases for Acidobacteria, Proteobacteria, and Verrucomicrobia [14]. However, despite these taxonomic differences, we did not identify significant predicted functional differences by depth. When we turned to specifically correlating the abundance of Group C to OTUs from the metagenome data, a large number of OTUs with positive or negative correlations were identified (Additional file 8: Dataset S7). At a broad level, the environmental data of the positively correlating taxa are consistent with Group C associating with other soil bacteria. While this provides scope for investigating specific taxa that may help shape the niche that Group C occupy, determining which taxa are important and the nature of their interactions will require a dedicated effort.
For the negatively correlating cohort, they tend to represent isolates from non-soil environments (Additional file 8: Dataset S7) and may therefore represent non-indigenous microorganisms that have been introduced. The permafrost samples were obtained from an “upland polygonal terrain in proximity to the McGill Arctic Research Station at Expedition Fjord (79°24’57"N, 90°45’46"W)” [13]. The prevalence of negatively correlating OTUs matching to Proteobacteria isolated from sea water may reflect aeolian carriage from Expedition Fjord, which is located ~8 km from the Research Station. As the samples were obtained for a simulated permafrost-thaw experiment [52], the negatively correlating OTUs may also reflect environmental disturbance.
Conclusions
Our study commenced with the analysis of genome sequences of new Group C Arthrobacter isolated from the Tibetan Plateau and progressed through to a rationalization of Group C abundance in global metagenomes. Group C was clearly distinguished from Group A Arthrobacter by possessing genomic signatures consistent with its representation in PA environments and an ability to grow faster when cultivated at −1°C. Assessment of available metagenome data points to the Group C traits as being more relevant to cold niches rather than competitiveness across global permafrost or cold soil environments. The challenge in being able to define the specific niche parameters enabling Group C Arthrobacter to be relatively competitive illustrates the inherent difficulties associated with defining “cause and effect” for explaining “why” microorganisms reside in the environments in which they are found, that is, the characteristics of the ecological niches that define microbiome structure [1]. Without knowing the specific effectors, the ability to understand and predict responses to environmental changes is greatly compromised [5, 7, 55, 56]. Establishing long-term data records that include comprehensive metadata associated with monitoring sites, including metadata for each biological sample, will be essential for learning how to link environmental parameters to microbial processes. In a study of sulfate reduction in Arctic marine sediments, growth yield was reasoned to be the most relevant factor for determining the competitiveness of sulfate-reducing bacteria in permanently cold marine sediments [54]. These findings illustrate that for cold environments, linking genomic and metagenomic data to measurements of metabolic rates, growth rates, and growth yields will undoubtedly help to clarify how specific microbial processes and associated taxa are influenced by environmental temperature.
While the characteristics that define the Group C niche are still to be defined, at sites where Group C Arthrobacter are relatively abundant, they may have value as a biomarker for monitoring the stability of those locations. Moreover, Group A Arthrobacter may serve as a more broadly useful biomarker of soil microbial communities. Group A exhibited high relative abundance across NPA metagenomes and relatively low abundance across PA metagenomes. As the data indicate environmental temperature exerts a broad, strong influence on Group A Arthrobacter, we predict that environmental warming will generally increase the relative abundance of Group A. Similar influences of environmental temperature have been described for the marine SAR11 clade, including the predicted displacement of polar specialists by phylotypes from warmer latitudes [57]. Depending on how strongly the environmental factors other than temperature select for Group C in permafrost, the apparent broad influence of temperature on Group A suggests it will displace Group C from the niches in which it is currently relatively competitive.
Methods
Arthrobacter isolation and genome sequencing
Sampling and isolation of Arthrobacter from lakes, glaciers, and a wetland on the Tibetan Plateau was performed based on procedures previously described [26, 27, 58, 59], and information associated with sampling and isolation is provided in Additional file 1: Table S1. Briefly, surface water samples from lakes Dawa Tso, Gomang Tso, Peng Tso, Ranwu, Sumzhi Tso, Yamdrok Tso, and Zigetangcuo were collected during the 2012 summer fieldwork based on procedures previously described [58]. All water samples were collected in sterile 250-mL Nalgene bottles and stored in the field at 4 °C. After transport at 4 °C to the Institute of Tibetan Plateau Research-Lhasa, 15% glycerol (v/v) was added and the samples were stored at −20 °C prior to transport and storage of samples at −20 °C at the Institute of Tibetan Plateau Research-Beijing. For glacier samples, 12-cm-diameter ice cores were drilled in Noijinkangsang (August 2007; 33-m-long ice core) and Ulugh Muztagh (May 2012; 164-m-long ice core) glaciers. The ice cores were cut into sub-sections with intervals of 5–10 cm using a bandsaw in a walk-in-freezer (−20 °C). Ice on the surface of the samples (1 cm thick) was chipped off using a sterilized blade, the inner cores were rinsed with cold ethanol (95%), followed by cold, triple-autoclaved, double-distilled water. The frozen lake water and the ice core samples were placed in autoclaved containers and melted slowly at 4 °C before being used for cultivation attempts. A volume of 200 μL of thawed water of each sample was placed directly onto R2A solid medium for cultivation.
Soil cores at 0–5-cm soil depth were collected from the Madoi wetland in August 2011 [59]. Soil samples were placed in a box with ice packs during transportation and were stored in the laboratory at 4 °C. The soil samples were suspended in triple-autoclaved, double-distilled water (m/v, 1:10), incubated statically at room temperature for 2 h, and 200 μL of supernatant was dispensed directly onto R2A solid medium for cultivation.
All cultivation on R2A solid medium (lake, glacier, and wetland samples) was performed at the Institute of Tibetan Plateau Research-Beijing in incubators at temperatures ranging from 4 to 24 °C for a period ranging from 1 week to 2 months (Additional file 1: Table S1). Colonies were quadrant-streaked several times for purification, and purity was assessed using microscopy.
Genomic DNA was extracted from isolates using a TIANamp Bacteria DNA Kit (Tiangen, Beijing) following the manufacturer’s instructions. The 16S rRNA genes were amplified with the universal bacterial primers 27F (5′AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′CGGTTACCTTGTTACGACTT-3′) and the amplification products were sequenced at Boai Yonghua (Beijing) on an ABI PRISM 3730xl sequencer. The taxonomy of the isolates was determined by aligning the 16S rRNA gene sequences against the NCBI-nr nucleotide database using blastn (Blast+ v2.9.0). The 16 Arthrobacter isolates (Additional file 1: Table S1) were deposited in the China General Microbiological Culture Collection Center (CGMCC) with accession numbers: CGMCC 1.16187-1.16198, 1.16223, and 1.16312.
Using genomic DNA (extracted as described above) for the 16 isolates, paired-end libraries with an insert size of 500 bp were constructed and sequenced using an Illumina Hiseq 2000 platform. Prior to de novo sequence assembly, low-quality reads were filtered out using Fastp with default options [60]. Filtered sequencing reads were subjected to assembly using SPAdes v3.11.1 with default options [61]. The assembled genome sequences were deposited in DDBJ/ENA/GenBank under the BioProject PRJNA421662.
For genomic analyses, three of the 16 isolates were excluded due to genome quality or dereplication criteria (also see the “Preparation of Arthrobacter genomes for analysis” section below and the “Phylogenomics” section in the “Results” section). All 13 isolates used for the study represented unique sites (lakes, glaciers, or wetland) or specific location of an individual lake (Zigetangcuo) or depth of a glacier core (Ulugh Muztagh) (Additional file 1: Table S1).
Growth temperature response
Three replicates of each Arthrobacter were grown in 100 mL of R2A broth in 150-mL flasks at 25, 5, and −1°C for up to 10 days. The optical density was measured at 600 nm (OD600) using a Microplate Reader (MD, SpectraMax M5) by transferring 200 μL of the culture into microwells. OD600 measurements were taken every 24 h for cultivation at −1 °C and 5 °C, and every 12 h for cultivation at 25 °C. For cultivation at −1°C, flasks were placed in ice produced by an ice maker (TKKY, FM40) with flasks placed in a ~ 4°C refrigerator and ice replaced every 12 h. Growth at 5 and 25°C was performed using a constant-temperature incubator as described previously [62]. All cultures were grown statically, with flasks swirled to resuspend biomass prior to recording OD600. Arthrobacter used for growth temperature profiles were as follows: Group A: A. luteolus, A. globiformis, and A. subterraneus; Group B: Arthrobacter sp. 4R501, Arthrobacter sp. 9E14, and Arthrobacter sp. 08Y14; and Group C: A. alpinus, Arthrobacter sp. A3, and Arthrobacter sp. N199823.
Preparation of Arthrobacter genomes for analysis
As the taxonomic assignment of the genus Arthrobacter is not consistent, in August 2018, all genome sequences with the taxonomy identifier “Arthrobacter” or “Micrococcaceae” were retrieved from GenBank, providing a total of 427 genomes including the 16 new Tibetan Plateau genomes. The completeness of each genome was calculated using CheckM v1.0.7 with default options [63]. Genomes composed of > 300 contigs, with an N50 of < 20 kb, completeness of < 95%, and contamination > 5% were removed. Genomes were dereplicated to remove genomes with an AAI ≥ 99.5%. AAI values were calculated using CompareM with default options (https://github.com/dparks1134/CompareM). ANI was calculated using the ANI calculator (http://enve-omics.ce.gatech.edu/ani/). A total of 210 genomes met quality requirements, which included 13 of the 16 new Tibetan Plateau genomes (Additional file 2: Dataset S1). Gene families were clustered using FastOrtho software (--pv_cutoff 1-e5 --pi_cutoff 70 --pmatch_cutoff 70) (http://enews.patricbrc.org/fastortho/) with the cutoff values set according to Parks et al. [35]. A gene family matrix was produced using custom PERL scripts, and non-functional-based group-specific genes were calculated based on this matrix. The annotation of genes was standardized by annotating all genomes using RAST (Rapid Annotation using Subsystem Technology) [64] and PROKKA [65].
Phylogenetic and genomic analyses
For phylogenomic clustering, Cellulomonas carbonis T26 and C. fimi ATCC 484 were chosen as the outgroup as they are close relatives of Micrococcaceae [66], and species that are closely related to the in-group are more suitable for phylogenetic reconstruction than distantly related species [67]. A maximum likelihood phylogenomic tree was constructed using PhyloPhlAn2 with default options [68]. A Bayesian tree was constructed using MPI Mrbayes v3.2 (prset aamodelpr = mixed, mcmc nchains = 16, ngen = 300,000, and leaving other parameter values as default) [69]. The F measure (harmonic mean of precision) provides a metric for determining if taxa are operationally monophyletic (F measure ≥ 0.95) [35] and was calculated as F = 2 × ((precision × recall)/(precision + recall)). The genome-wide amino acid composition was calculated using CompareM with the function aa_usage. The stability curves of proteins were predicted by SCooP [37] using the PDB (Protein Data Bank) files modeled by SWISS-MODEL [70]. The stability equations of the same protein from different hosts were visualized and smoothed using ggplot2 v3.2.1 [71]. The stability curves were analyses for 180 single-copy genes that were shared by most genomes; a small number of genomes had multiple copies of genes, and up to three genomes were permitted to have the absence of the gene in order to account for the use of unclosed genomes (99 of the 106). After retrieval from SWISS-MODEL of all possible PDB files matching the candidate genes, a total of 17,339 stability equations were generated (Additional file 3: Dataset S2). Ordination and statistical analyses, including three-dimensional nonmetric multidimensional scaling and gene enrichment analyses, were performed with R v3.3.3 and Origin v9.5. For comparisons between Group A, B, and C Arthrobacter, group-specific genes or functions were defined as being present in 95% of the target group (e.g., Group C) genomes and absent in 95% of each of the other group(s) (e.g., Group A) genomes. Group-specific genes were identified (Group A, 74 genomes, 16,149 specific genes; Group B, 22 genomes, 4675 specific genes; Group C, 10 genomes, 969 specific genes; Additional file 6: Dataset S5) and normalized to account for the different number of genomes used for each group. To account for differences in gene content between Arthrobacter, comparisons were calculated relative to the total Arthrobacter gene complement from all 106 Arthrobacter genomes (referred to as the Arthrobacter pan genome). Gene copy number was calculated as the average number of the gene for each genome in a group (e.g., Group C), with gene loss or gain calculated from the average copy number for the groups (e.g., Group C vs Group A). To assess the bias of amino acid composition of different functional classes of proteins, genes were assigned to functional categories (assigned by RAST) and total amino acid composition for all proteins from the functional category was compared between groups (e.g., Group C vs Group A). Similarity was measured by Bray-Curtis distance with larger Bray-Curtis distances denoting stronger bias, possibly indicative of selection pressure [41]. The functional potential of groups was also compared using enrichment analysis [41]. Briefly, the presence or absence of KEGG Ortholog (KO) groups in genomes and metagenomes (see the “Collection and analysis of metagenomes” section) was assessed [41], and non-parametric one-way ANOVA was used to identify differentially abundant categories using R [72].
Collection and analysis of metagenomes
Assembled metagenomes were downloaded from IMG (https://genome.jgi.doe.gov/portal/). Classification into PA or NPA environments were made using metadata associated with metagenomes, supplemented by Köppen-Geiger climate classifications (to define temperate and tropical regions) using ArcGIS location data [73]. Analyses were initially performed using 639 metagenomes from environments with MAT ranging from −24 to 28 °C, representing PA (n = 196, gene count = 183 million), temperate (n = 243, gene count = 190 million), and tropical (n = 200, gene count = 841 million) zones (Additional file 7: Dataset S6). Subsequently, all additional (334) available (May 2020) unique assembled permafrost metagenomes were analyzed. Analyses assessed the relative abundance of each Arthrobacter group (A, B, and C) using group-specific genes (see the “Phylogenetic and genomic analyses” section) by performing a local alignment search against the metagenomes using DIAMOND v0.9.24 with the arguments --outfmt 6, --query-cover 70, --id 70, --evalue 1e-5, and leaving others as default [74]. One-way ANCOVA was used to assess statistical differences between regression lines for groups of metagenomes [75] using the data import webform for k = 2 at http://vassarstats.net/vsancova.html. Correlation analyses were performed between Arthrobacter groups and other members of the microbial community from 76 metagenomes derived from a simulated permafrost-thaw experiment [14]. OTUs were assigned from IMG phylodist matches (which are based on the top taxon in the IMG isolate database) by clustering the IMG phylodist matches at the genus level; < 4% of OTUs had < 35% identity. The raw abundance of all OTUs was determined, with 956 meeting the criteria of average abundance ≥ 2, and detection in at least 56 of the 76 metagenomes. The 956 OTUs were used to construct a correlation matrix using SparCC [76] implemented in python3 with default parameters (20 iterations). One hundred simulated datasets were created by random shuffling of the original input with replacement, and their correlation matrices were constructed in the same way. The simulated datasets were used to calculate the one- and two-sided pseudo p-values. The selected threshold for strong correlations was > | 0.5 |. To assess if depth, incubation time, treatment group, sample core, or physicochemical data explained the variation in species composition across permafrost sites, a generalized linear latent variable model was employed as implemented in the R package gllvm [77]. All custom scripts are available at https://github.com/environmental-genomes/Arthro.
Supplementary Information
Additional file 1: Tibetan Plateau sampling sites, Arthrobacter phylogenetic, genomic, functional and metagenomic analyses. Supplementary figures. Figure S1. Tibetan Plateau sampling locations. Figure S2. Micrococcaceae phylogenomic trees. Figure S3. Comparison of Arthrobacter genomic characteristics. Figure S4. Comparison of Arthrobacter genome-wide amino acid composition. Figure S5. Correlation between the content of an amino acid and genome G+C content. Figure S6. Protein stability predictions. Figure S7. Enrichment analysis of Arthrobacter functional potential. Supplementary tables. Table S1. Sampling and isolation information associated with Tibetan Plateau Arthrobacter isolates. Table S2. Depth distribution of the 11 Axel Heiberg Island metagenomes enriched in Group C Arthrobacter.
Additional file 2: Dataset S1. Micrococcaceae and Arthrobacter genomes used in this study.
Additional file 3: Dataset S2. Proteins and stability equations used for stability predictions.
Additional file 4: Dataset S3. Data used for enrichment analyses of functional categories.
Additional file 5: Dataset S4. Gene copy number data.
Additional file 6: Dataset S5. Protein sequences of Arthrobacter group-specific genes.
Additional file 7: Dataset S6. Metagenomes used in this study.
Additional file 8: Dataset S7. Correlation of the abundance of Arthrobacter with taxa from Axel Heiberg Island metagenomes.
Acknowledgements
We thank Qilong Qin, Joshua N. Hamm, Pratibha Panwar, Sten Anslan, Saleh Rahimlou, Ping Ren, Fei Liu, Weizhi Song, Wei Zhu, Zhihao Zhang, Wenqiang Wang, and Yongcui Deng for their valuable input related to data analyses or providing maps or images of the sampling sites, and Maggie Lau, Lyle Whyte, and Tullis Onstott for information related to the Axel Heiberg Island site. Metagenome sequence data used in the construction of Figure 5 are produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community. We acknowledge the efforts of all the scientists involved in enabling the data to be generated and thank all PIs for providing permission to utilize data.
Authors’ contributions
YL, RC, and LS designed the study; YL, BX, and NW collected field samples; YZ and QL provided type strains; LS and FW performed the experiments; LS and MAA analyzed the data; RC, LS, TJW, and MAA interpreted the data and wrote the paper. The authors read and approved the final manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (Grant Nos. 91851207 and 41701085), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK0503), and the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDA20050101). The Australian contingent was supported by funding from the Australian Research Council (DP150100244). Liang Shen was funded by the China Scholarship Council (Grant No. 201804910177).
Availability of data and materials
The 16 newly isolated Arthrobacter were deposited in the China General Microbiological Culture Collection Center (CGMCC) with accession numbers: CGMCC 1.16187-1.16198, 1.16223, and 1.16312. The assembled genome sequences for newly isolated Arthrobacter were deposited in DDBJ/ENA/GenBank under the BioProject PRJNA421662. All custom scripts are available at https://github.com/environmental-genomes/Arthro.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongqin Liu, Email: yqliu@itpcas.ac.cn.
Ricardo Cavicchioli, Email: r.cavicchioli@unsw.edu.au.
References
- 1.Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8. [DOI] [PMC free article] [PubMed]
- 2.Grote M. Petri dish versus Winogradsky column: a longue durée perspective on purity and diversity in microbiology, 1880s–1980s. Hist Philos Life Sci. 2018;40:113. doi: 10.1007/s40656-017-0175-9. [DOI] [PubMed] [Google Scholar]
- 3.Colman DR, Poudel S, Stamps BW, Boyd ES, Spear JR. The deep, hot biosphere: twenty-five years of retrospection. Proc Natl Acad Sci U S A. 2017;114(27):6895–6903. doi: 10.1073/pnas.1701266114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541(7637):353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 5.Cavicchioli R, Ripple WJ, Timmis KN, Azam F, Bakken LR, Baylis M, Behrenfeld MJ, Boetius A, Boyd PW, Classen AT, Crowther TW, Danovaro R, Foreman CM, Huisman J, Hutchins DA, Jansson JK, Karl DM, Koskella B, Mark Welch DB, Martiny JBH, Moran MA, Orphan VJ, Reay DS, Remais JV, Rich VI, Singh BK, Stein LY, Stewart FJ, Sullivan MB, van Oppen MJH, Weaver SC, Webb EA, Webster NS. Scientists’ warning to humanity: microorganisms and climate change. Nat Rev Microbiol. 2019;17(9):569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timmis K, Cavicchioli R, Garcia JL, Nogales B, Chavarria M, Stein L, et al. The urgent need for microbiology literacy in society. Environ Microbiol. 2019;21(5):1513–1528. doi: 10.1111/1462-2920.14611. [DOI] [PubMed] [Google Scholar]
- 7.Edwards A, Cameron KA, Cook JM, Debbonaire AR, Furness E, Hay MC, et al. Microbial genomics amidst the Arctic crisis. Microb Genom. 2020;6. [DOI] [PMC free article] [PubMed]
- 8.Chen LX, Anantharaman K, Shaiber A, Eren AM, Banfield JF. Accurate and complete genomes from metagenomes. Genome Res. 2020;30(3):315–333. doi: 10.1101/gr.258640.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nkongolo KK, Narendrula-Kotha R. Advances in monitoring soil microbial community dynamic and function. J Appl Genet. 2020;61(2):249–263. doi: 10.1007/s13353-020-00549-5. [DOI] [PubMed] [Google Scholar]
- 10.Cavicchioli R. Microbial ecology of Antarctic aquatic systems. Nat Rev Microbiol. 2015;13(11):691–706. doi: 10.1038/nrmicro3549. [DOI] [PubMed] [Google Scholar]
- 11.Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M, Cavicchioli R, Murray AE. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012;6(10):1901–1915. doi: 10.1038/ismej.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panwar P, Allen MA, Williams TJ, Hancock AM, Brazendale S, Bevington J, Roux S, Páez-Espino D, Nayfach S, Berg M, Schulz F, Chen IMA, Huntemann M, Shapiro N, Kyrpides NC, Woyke T, Eloe-Fadrosh EA, Cavicchioli R. Influence of the polar light cycle on seasonal dynamics of an Antarctic lake microbial community. Microbiome. 2020;8(1):116. doi: 10.1186/s40168-020-00889-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau MCY, Stackhouse BT, Layton AC, Chauhan A, Vishnivetskaya TA, Chourey K, Ronholm J, Mykytczuk NCS, Bennett PC, Lamarche-Gagnon G, Burton N, Pollard WH, Omelon CR, Medvigy DM, Hettich RL, Pfiffner SM, Whyte LG, Onstott TC. An active atmospheric methane sink in high Arctic mineral cryosols. ISME J. 2015;9(8):1880–1891. doi: 10.1038/ismej.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stackhouse BT, Vishnivetskaya TA, Layton A, Chauhan A, Pfiffner S, Mykytczuk NC, Sanders R, Whyte LG, Hedin L, Saad N, Myneni S, Onstott TC. Effects of simulated spring thaw of permafrost from mineral cryosol on CO2 emissions and atmospheric CH4 uptake. J Geophys Res Biogeosci. 2015;120(9):1764–1784. doi: 10.1002/2015JG003004. [DOI] [Google Scholar]
- 15.Conn H, Dimmick I. Soil bacteria similar in morphology to Mycobacterium and Corynebacterium. J Bacteriol. 1947;54(3):291–303. doi: 10.1128/JB.54.3.291-303.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cacciari I, Lippi D. Arthrobacters: successful arid soil bacteria: a review. Arid Soil Res Rehabil. 1987;1(1):1–30. doi: 10.1080/15324988709381125. [DOI] [Google Scholar]
- 17.Niewerth H, Schuldes J, Parschat K, Kiefer P, Vorholt JA, Daniel R, Fetzner S. Complete genome sequence and metabolic potential of the quinaldine-degrading bacterium Arthrobacter sp. Rue61a. BMC Genomics. 2012;13(1):534. doi: 10.1186/1471-2164-13-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busse HJ, Wieser M, et al. Arthrobacter. In: Trujillo ME, Dedysh S, DeVos P, Hedlund B, Kämpfer P, Rainey FA, et al., editors. Bergey’s manual of systematics of Archaea and Bacteria. Hoboken, NJ: Wiley; 2018. pp. 1–43. [Google Scholar]
- 19.Unell M, Nordin K, Jernberg C, Stenstrom J, Jansson JK. Degradation of mixtures of phenolic compounds by Arthrobacter chlorophenolicus A6. Biodegradation. 2008;19(4):495–505. doi: 10.1007/s10532-007-9154-2. [DOI] [PubMed] [Google Scholar]
- 20.Boylen CW. Survival of Arthrobacter crystallopoietes during prolonged periods of extreme desiccation. J Bacteriol. 1973;113(1):33–37. doi: 10.1128/JB.113.1.33-37.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T, Mukouyama M, Sakano K, Tani Y. Degradation of a sodium acrylate oligomer by an Arthrobacter sp. Appl Environ Microbiol. 1993;59(5):1555–1559. doi: 10.1128/AEM.59.5.1555-1559.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dsouza M, Taylor MW, Turner SJ, Aislabie J. Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genomics. 2015;16(1):36. doi: 10.1186/s12864-015-1220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gounot A. Effects of temperature on the growth of psychrophilic bacteria from glaciers. Can J Microbiol. 1976;22(6):839–846. doi: 10.1139/m76-121. [DOI] [PubMed] [Google Scholar]
- 24.Feller G. Psychrophilic enzymes: from folding to function and biotechnology. Scientifica. 2013;2013:512840. doi: 10.1155/2013/512840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X-M, Jiang Y, Li Y-T, Zhang H-H, Li J, Chen X, Zhao Q, Zhao J, Si J, Lin ZW, Zhang H, Dyson P, An LZ. Regulation of expression of trehalose-6-phosphate synthase during cold shock in Arthrobacter strain A3. Extremophiles. 2011;15(4):499–508. doi: 10.1007/s00792-011-0380-5. [DOI] [PubMed] [Google Scholar]
- 26.Shen L, Liu YQ, Wang N, Jiao NZ, Xu BQ, Liu XB. Variation with depth of the abundance, diversity and pigmentation of culturable bacteria in a deep ice core from the Yuzhufeng Glacier, Tibetan Plateau. Extremophiles. 2018;22(1):29–38. doi: 10.1007/s00792-017-0973-8. [DOI] [PubMed] [Google Scholar]
- 27.Liu YQ, Priscu JC, Yao TD, Vick-Majors TJ, Michaud AB, Sheng L. Culturable bacteria isolated from seven high-altitude ice cores on the Tibetan Plateau. J Glaciol. 2019;65(249):29–38. doi: 10.1017/jog.2018.86. [DOI] [Google Scholar]
- 28.Margesin R, Feller G. Biotechnological applications of psychrophiles. Environ Technol. 2010;31(8-9):835–844. doi: 10.1080/09593331003663328. [DOI] [PubMed] [Google Scholar]
- 29.Cavicchioli R, Charlton T, Ertan H, Mohd Omar S, Siddiqui KS, Williams TJ. Biotechnological uses of enzymes from psychrophiles. Microb Biotechnol. 2011;4(4):449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anesio AM, Laybourn-Parry J. Glaciers and ice sheets as a biome. Trends Ecol Evol. 2012;27(4):219–225. doi: 10.1016/j.tree.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Siddiqui KS, Williams TJ, Wilkins D, Yau S, Allen MA, Brown MV, Lauro FM, Cavicchioli R. Psychrophiles. Annu Rev Earth Planet Sci. 2013;41(1):87–115. doi: 10.1146/annurev-earth-040610-133514. [DOI] [Google Scholar]
- 32.Boetius A, Anesio MA, Deming WJ, Mikucki AJ, Rapp ZJ. Microbial ecology of the cryosphere: sea ice and glacial habitats. Nat Rev Microbiol. 2015;13(11):677–690. doi: 10.1038/nrmicro3522. [DOI] [PubMed] [Google Scholar]
- 33.Cavicchioli R. On the concept of a psychrophile. ISME J. 2016;10(4):793–795. doi: 10.1038/ismej.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins T, Margesin R. Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Appl Microbiol Biotechnol. 2019;103(7):2857–2871. doi: 10.1007/s00253-019-09659-5. [DOI] [PubMed] [Google Scholar]
- 35.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36(10):996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 36.Loveland J, Gutshall K, Kasmir J, Prema P, Brenchley JE. Characterization of psychrotrophic microorganisms producing beta-galactosidase activities. Appl Environ Microbiol. 1994;60(1):12–18. doi: 10.1128/AEM.60.1.12-18.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pucci F, Kwasigroch JM, Rooman M. SCooP: an accurate and fast predictor of protein stability curves as a function of temperature. Bioinformatics. 2017;33(21):3415–3422. doi: 10.1093/bioinformatics/btx417. [DOI] [PubMed] [Google Scholar]
- 38.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311(5760):496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 39.Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J Bacteriol. 2008;190(5):1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS, De Francisci D, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 2009;3(9):1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 41.Bai Y, Muller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528(7582):364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 42.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72(3):471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bzymek KP, Newton GL, Ta P, Fahey RC. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J Bacteriol. 2007;189(19):6796–6805. doi: 10.1128/JB.00644-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang XJ, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci U S A. 2005;102(31):10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams TJ, Lauro FM, Ertan H, Burg DW, Poljak A, Raftery MJ, Cavicchioli R. Defining the response of a microorganism to temperatures that span its complete growth temperature range (-2 °C to 28 °C) using multiplex quantitative proteomics. Environ Microbiol. 2011;13(8):2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- 46.Dsouza M, Taylor MW, Turner SJ, Aislabie J. Genome-based comparative analyses of Antarctic and temperate species of Paenibacillus. PLoS One. 2014;9(10):e108009. doi: 10.1371/journal.pone.0108009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goordial J, Raymond-Bouchard I, Zolotarov Y, de Bethencourt L, Ronholm J, Shapiro N, et al. Cold adaptive traits revealed by comparative genomic analysis of the eurypsychrophile Rhodococcus sp. JG3 isolated from high elevation McMurdo Dry Valley permafrost, Antarctica. FEMS Microbiol Ecol. 2016;92:fiv154. doi: 10.1093/femsec/fiw154. [DOI] [PubMed] [Google Scholar]
- 48.Mackelprang R, Burkert A, Haw M, Mahendrarajah T, Conaway CH, Douglas TA, Waldrop MP. Microbial survival strategies in ancient permafrost: insights from metagenomics. ISME J. 2017;11(10):2305–2318. doi: 10.1038/ismej.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer JL, Abeles RH. The mechanism of action of S-adenosylhomocysteinase. J Biol Chem. 1979;254(4):1217–1226. doi: 10.1016/S0021-9258(17)34190-X. [DOI] [PubMed] [Google Scholar]
- 50.Sah S, Aluri S, Rex K, Varshney U. One-carbon metabolic pathway rewiring in Escherichia coli reveals an evolutionary advantage of 10-formyltetrahydrofolate synthetase (Fhs) in survival under hypoxia. J Bacteriol. 2015;197(4):717–726. doi: 10.1128/JB.02365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2(11):1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 52.Chauhan A, Layton AC, Vishnivetskaya TA, Williams D, Pfiffner SM, Rekepalli B, et al. Metagenomes from thawing low-soil-organic-carbon mineral cryosols and permafrost of the Canadian high arctic. Genome Announc. 2014;2:e01217–e01214. doi: 10.1128/genomeA.01217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu Rev Biochem. 2006;75(1):403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 54.Scholze C, Jørgensen BB, Røy H. Psychrophilic properties of sulfate-reducing bacteria in Arctic marine sediments. Limnol Oceanogr. 2020. 10.1002/lno.11586.
- 55.Webster NS, Wagner M, Negri AP. Microbial conservation in the Anthropocene. Environ Microbiol. 2018;20(6):1925–1928. doi: 10.1111/1462-2920.14124. [DOI] [PubMed] [Google Scholar]
- 56.Cavicchioli R. A vision for a ‘microbcentric’ future. Microb Biotechnol. 2019;12(1):26–29. doi: 10.1111/1751-7915.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T, et al. Global biogeography of SAR11 marine bacteria. Mol Syst Biol. 2012;8(1):595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Yao T, Jiao N, Zhu L, Hu A, Liu X, Gao J, Chen ZQ. Salinity impact on bacterial community composition in five high-altitude lakes from the Tibetan Plateau, Western China. Geomicrobiol J. 2013;30(5):462–469. doi: 10.1080/01490451.2012.710709. [DOI] [Google Scholar]
- 59.Deng Y, Cui X, Hernandez M, Dumont MG. Microbial diversity in hummock and hollow soils of three wetlands on the Qinghai-Tibetan Plateau revealed by 16S rRNA pyrosequencing. PLoS One. 2014;9(7):e103115. doi: 10.1371/journal.pone.0103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen L, Liu YQ, Xu BQ, Wang NL, Zhao HB, Liu XB, Liu F. Comparative genomic analysis reveals the environmental impacts on two Arcticibacter strains including sixteen Sphingobacteriaceae species. Sci Rep. 2017;7(1):2055. doi: 10.1038/s41598-017-02191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42(D1):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 66.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rosselló-Móra R. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31(4):241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Yang ZH. Computational molecular evolution. Great Britain: Oxford University Press; 2006. [Google Scholar]
- 68.Segata N, Bornigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4(1):2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
- 72.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 73.Peel MC, Finlayson BL, Mcmahon TA. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci. 2007;11:259–263. doi: 10.5194/hess-11-1633-2007. [DOI] [Google Scholar]
- 74.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 75.McDonald JH. Handbook of biological statistics. 3. Baltimore, Maryland: Sparky House Publishing; 2014. [Google Scholar]
- 76.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Niku J, Hui FKC, Taskinen S, Warton DI. gllvm: fast analysis of multivariate abundance data with generalized linear latent variable models in R. Methods Ecol Evol. 2019;10:2173–2182. doi: 10.1111/2041-210X.13303. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Tibetan Plateau sampling sites, Arthrobacter phylogenetic, genomic, functional and metagenomic analyses. Supplementary figures. Figure S1. Tibetan Plateau sampling locations. Figure S2. Micrococcaceae phylogenomic trees. Figure S3. Comparison of Arthrobacter genomic characteristics. Figure S4. Comparison of Arthrobacter genome-wide amino acid composition. Figure S5. Correlation between the content of an amino acid and genome G+C content. Figure S6. Protein stability predictions. Figure S7. Enrichment analysis of Arthrobacter functional potential. Supplementary tables. Table S1. Sampling and isolation information associated with Tibetan Plateau Arthrobacter isolates. Table S2. Depth distribution of the 11 Axel Heiberg Island metagenomes enriched in Group C Arthrobacter.
Additional file 2: Dataset S1. Micrococcaceae and Arthrobacter genomes used in this study.
Additional file 3: Dataset S2. Proteins and stability equations used for stability predictions.
Additional file 4: Dataset S3. Data used for enrichment analyses of functional categories.
Additional file 5: Dataset S4. Gene copy number data.
Additional file 6: Dataset S5. Protein sequences of Arthrobacter group-specific genes.
Additional file 7: Dataset S6. Metagenomes used in this study.
Additional file 8: Dataset S7. Correlation of the abundance of Arthrobacter with taxa from Axel Heiberg Island metagenomes.
Data Availability Statement
The 16 newly isolated Arthrobacter were deposited in the China General Microbiological Culture Collection Center (CGMCC) with accession numbers: CGMCC 1.16187-1.16198, 1.16223, and 1.16312. The assembled genome sequences for newly isolated Arthrobacter were deposited in DDBJ/ENA/GenBank under the BioProject PRJNA421662. All custom scripts are available at https://github.com/environmental-genomes/Arthro.