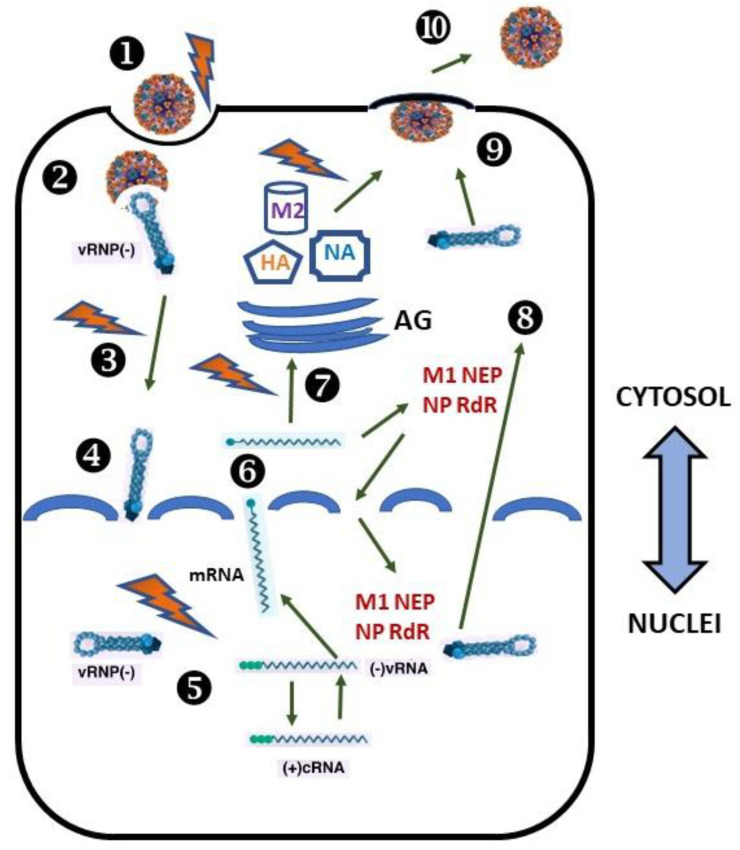
Figure 1.
Schematic representation of the important steps of the IAV life cycle. Recognition and attachment (Stage 1). Viral HA (orange) recognizes the terminal α2,6- or α2,3-linked NANA residues in glycan of the sialo-containing receptor on the plasma membrane and attaches to them. Entry (Stage 2). Virus entry to the host cell via endocytosis. Fusion of endosomal and viral membranes and release of viral genome (Stage 3). Once inside the acidic endosomal compartment and after the structural rearrangement of HA, the lipid bilayers of the virus and endosome fuse, and a pore is formed. Transfer to nuclei (Stage 4). Viral genome packed in vRNPs is released into the cytosol through the formed pore. Transcription, replication, and assembly (Stage 5). Later, they are transferred to the nuclei. Transcription and replication of the IAV genome occur at the genomic vRNA in the nuclei. Synthesis and maturation viral proteins (Stage 6 and 7). v-mRNAs are transported into the cytosol and translated on membrane-bound or free polyribosomes. Packaging of vRNPs (Stage 8). Viral proteins that are needed in virus assembly (M1, NP, NEP, and RNA-dependent RNA-polymerase subunits) return to the nucleus to be packaged as vRNPs. Later, they are exported to the cytosol and delivered to the plasma membrane. Budding and realizing virus particles (Stage 9 and 10). Envelope viral proteins (HA, NA, and M2) are synthesized on the rough ER and enter the cellular secretory pathway. Here, HA and NA co-translationally acquire the N-carbohydrate block, which completes their maturation in the Golgi. After that, they are transported via post-Golgi transport machinery to the PM and meet the vRNPs. Here, the budding and release of progeny viruses occur. Protein M1 assists in the formation of mature virions, using fragments of the lipid bilayer and some proteins of the host cell. The viral NA cleaves sialic acid from cell-surface glycoproteins and glycolipids, and virions are released.