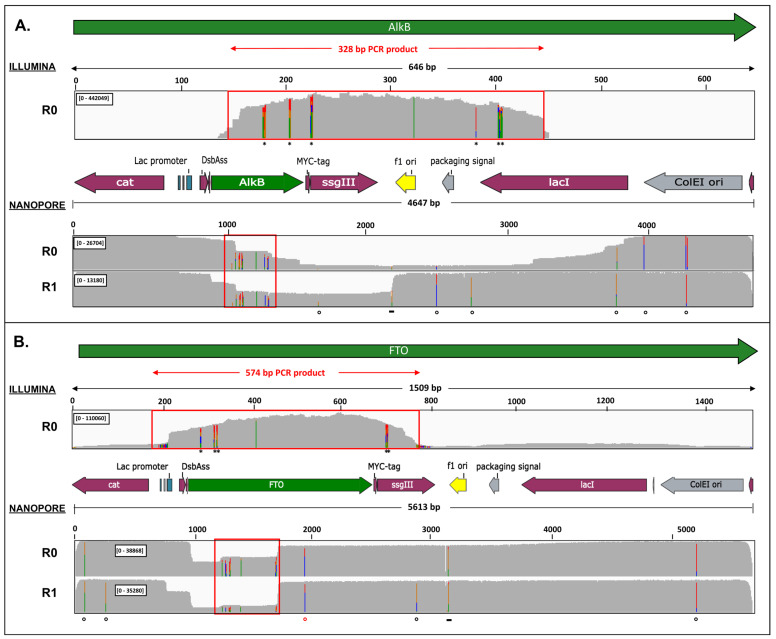
Figure 3.
Sequence coverage of Illumina and nanopore sequence reads of the AlkB (A) and FTO (B) libraries before (R0) and after one selection round (R1). Sequence coverage (gray scale) was obtained after Illumina read processing (quality/adapter trimming, pairing) of PCR-amplified, tagmented target regions (top part of each panel). Nanopore sequence coverage (bottom part of each panel) resulted from the alignment of quality-filtered nanopore reads of ScaI-linearized phagemids to the full display vector reference sequence. Asterisks (*,**) indicate the randomized alkB and FTO target codons. Open black dots represent recurrent substitutions which are assumed to have no functional effects. The open red dot represents a frequent ochre mutation at FTO codon 314. A recurrent short deletion (black bar (-)) is caused by erroneous read-out of a GC-rich region. The Illumina sequencing window is marked by a red box (shown in more detail in Figure S7). Functional features of pDST32-AlkB/FTO vectors are shown as horizontal arrows.