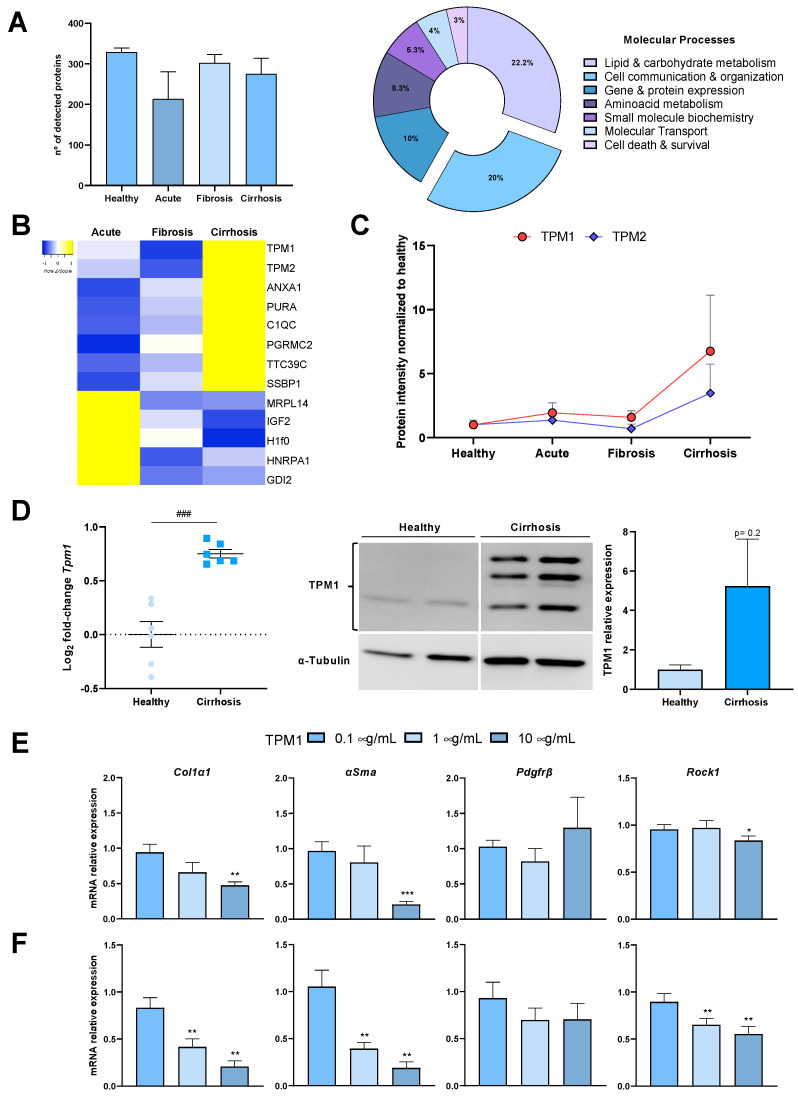
Figure 5.
LSECs extracellular vesicles proteomics during CLD progression. (A) Left, number of proteins detected in LSECs-derived small EVs at each stage of CLD progression. Right, cellular processes of deregulated proteins in cirrhotic LSECs EVs analysed by Ingenuity Pathway Analysis. (B) Heatmap of the top up- (yellow) and down-regulated (blue) proteins of LSECs EVs during CLD progression and, (C) graphical representation of Tpm1 & 2 gene expression in LSECs EVs across CLD. (D) Left, mRNA expression of Tpm1 in healthy and CCl4-cirrhotic LSECs. Values are represented in log2 fold-change. Right, western blot representative image of TPM1 in healthy and CCl4-cirrhotic LSECs, and corresponding quantification. (E) mRNA expression of depicted genes in in vitro activated HSCs and (F) primary isolated cirrhotic CCl4-HSCs exposed to different concentrations of recombinant TPM1 for 24 h. Red dot lines represent vehicle-treated cells. Results derive from n = 9 isolations (A,B), n = 9 independent samples (C), n = 6 independent isolations (D) and n = 3–5 independent experiments (E,F). ### p < 0.001 t test between cirrhotic and healthy group; * p < 0.05, ** p < 0.01 and *** p < 0.001, t test (Mann-Whitney U test for non-parametric variables) for differences between each group and the corresponding control group.