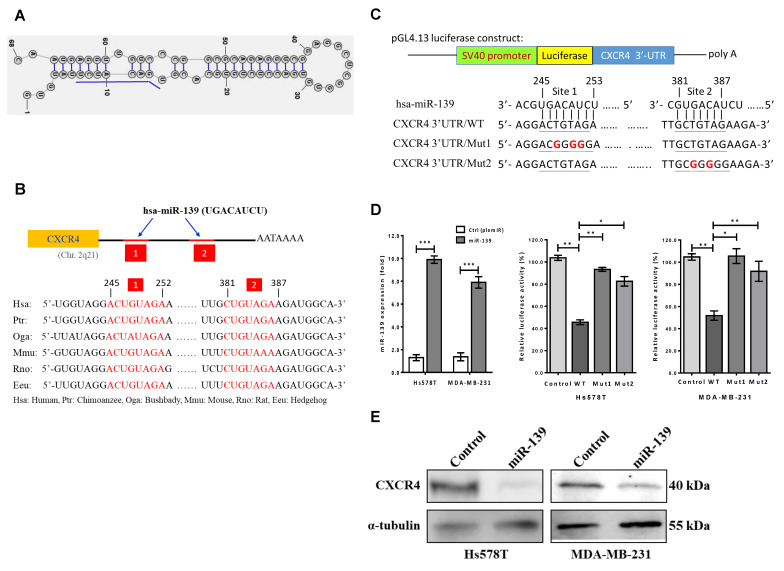
Figure 2.
CXCR4 as a direct target of hsa-miR-139-5p. (A) The stem-loop sequences hsa-miR-139-5p (miR-139) using RNAstructure, Version 6.2 and sequences in the mature miR-139 (underlined in blue) are shown. (B) Prediction of the targeting sequence for miR-139 within the 3′-UTR of CXCR mRNA. Two binding motifs in the 3′-UTR of CXCR4 gene for miR-139 (red) are shown across different vertebrate species. (C) Schematic representation of the luciferase reporter constructs showed a mismatch of the miR-139 binding sequence that are underlined at site 1 (CXCR4 3′-UTR/Mut1) and site 2 (CXCR4 3′-UTR/Mut2), respectively. (D) The dual-luciferase reporter (DLR™) assay was evaluated in breast cancer cell lines expressing the constructs designed in (C). Firefly luciferase activity was normalized to Renilla luciferase activity and compared with the cells overexpressing miR-139 or empty vector (control). Data are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01. (E) A significantly decreased expression of CXCR4 was observed in both invasive breast cancer cell lines transfected with miR-139. α-tubulin served as the loading control.