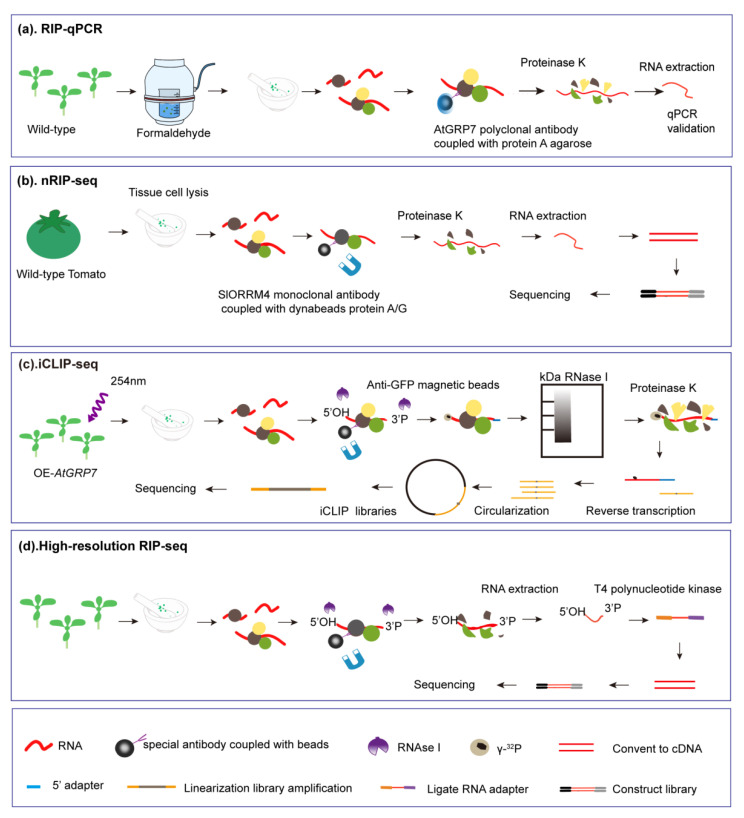
Figure 2.
Workflow of RIP-qPCR, native RIP-seq, iCLIP and high-resolution RIP-seq analyses of plants. (a) RIP with a AtGRP7-specific antibody followed by qPCR to identify the binding status of AtGRP7 at the FLC locus. Point mutation of R49Q (Arg to Gln at the 49th amino acid) in the RRM motif was used as a negative control. Arabidopsis seedling tissue was cross-linked by 0.5% (v/v) formaldehyde. After tissue lysis, immunoprecipitation was performed using anti-AtGRP7 antibodies coupled with protein A agarose. Then, the protein was degraded by proteinase K, and RNA was extracted by acidic phenol/chloroform. The percentage of RIP-enriched RNA relative to that of input sample was determined by qRT-PCR. (b) For native RIP, tomato fruit (36 days post-anthesis (DPA)) cells were directly lysed and immunoprecipitation was performed using anti-SlORRM4 antibodies coupled with dynabeads protein A/G and proteins were digested using proteinase K. RNA extraction was performed and analyzed by next-generation sequencing. The negative control was IP from slorrm4 mutant with anti-SlORRM4. (c) The GFP-tagged AtGRP7 was expressed in the grp7 mutant. Plant materials used for iCLIP were subjected to crosslinking with UV light 254 nm wavelength. After sample homogenization in liquid N2, a lysate was prepared and RNA–protein complexes were precipitated using specific antibodies coupled with magnetic beads. RNAs were fragmented by treatment with RNAse I and the fragments were radioactively labeled at the 5′ end only. After proteins were digested and RNAs were isolated, iCLIP sequencing libraries were prepared. In parallel, the negative control libraries were immunoprecipitated for GFP-only transgene plants and AtGRP7 R49Q-GFP plants. (d) For high-resolution RIP-seq, tissues extracts were pre-treated with RNAse I. Immunoprecipitation was then performed using a specific antibody that recognized the protein of interest. After the proteins were digested, the RNA was phosphorylated using a T4 polynucleotide kinase and processed for sequencing using a Small RNA-seq Kit.