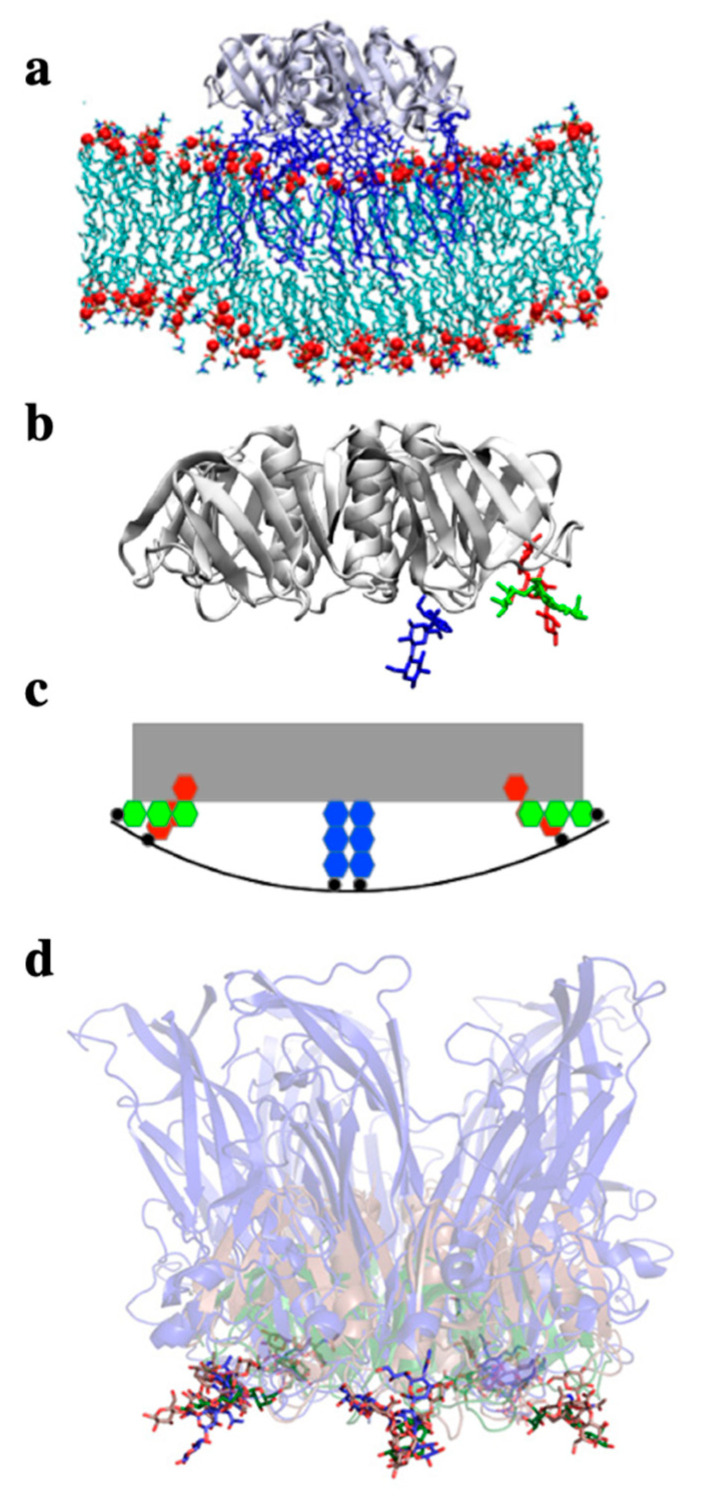
Figure 5.
Negative curvature induction based on the geometry of GSL binding sites on lectins. (a) All atom molecular dynamics simulation of the STxB homopentamer (gray ribbon structure, side view) on a membrane patch. Gb3 molecules in blue. Note the increment of spontaneous membrane curvature that is observed in silico due to the presence of STxB. (b) Representation (extracted from (a)) of STxB (side view) with three carbohydrates (blue, green and red). For simplicity, only the three Gb3 binding sites on one monomer are shown. Note that sites 1 (red) and 2 (green) are at the periphery of the protein, while site 3 (blue) faces down under the protein. (c) Schematic representation of the situation in (b) which illustrates that the membrane (black line) needs to bend up at the edges of STxB to plug the sugars of Gb3 receptor molecules (red and green) into the binding pockets of sites 1 and 2. This would then lead to the generation of negative membrane curvature. (d) Overlay of the structures of the following proteins in interaction with GSL receptor analogues: STxB in green with Gb3 (Reference [53]), CTxB in red with GM1 (Reference [54]), the capsid protein VP1 from SV40 in blue (Reference [55]). Note that despite the fact that these proteins do not have any sequence similarity, they fold such that the conserved GSL binding site 2 is presented with the same geometry in space (the sugars of the three structures overlay). It therefore seems likely that this fold was selected by convergent evolution for the same function: to generate negative membrane curvature for building endocytic sites for clathrin-independent endocytosis. Reproduced with permission from Toxins, Ref. [24].