Abstract
Simple Summary
Chromosome instability (CIN) is an increased rate where chromosome acquire alterations due to errors in cell division. CIN creates genetic and cytogenetic diversity and is a common feature in hematological malignancies such as acute myeloid leukemia (AML). Low to moderate levels of CIN seems to be well tolerated and can promote cancer proliferation, genetic diversity, and tumor evolution. However, high levels of CIN seems to be lethal, where enhancing CIN could improve AML treatment. However, little is known about CIN in AML. Our review focus on CIN studies in AML, their prognostic results, as well as the use of CIN as a therapeutic target in AML.
Abstract
Chromosomal instability (CIN), the increasing rate in which cells acquire new chromosomal alterations, is one of the hallmarks of cancer. Many studies highlighted CIN as an important mechanism in the origin, progression, and relapse of acute myeloid leukemia (AML). The ambivalent feature of CIN as a cancer-promoting or cancer-suppressing mechanism might explain the prognostic variability. The latter, however, is described in very few studies. This review highlights the important CIN mechanisms in AML, showing that CIN signatures can occur largely in all the three major AML types (de novo AML, secondary-AML, and therapy-related-AML). CIN features in AML could also be age-related and reflect the heterogeneity of the disease. Although most of these abnormalities show an adverse prognostic value, they also offer a strong new perspective on personalized therapy approaches, which goes beyond assessing CIN in vitro in patient tumor samples to predict prognosis. Current and emerging AML therapies are exploring CIN to improve AML treatment, which includes blocking CIN or increasing CIN beyond the limit threshold to induce cell death. We argue that the characterization of CIN features, not included yet in the routine diagnostic of AML patients, might provide a better stratification of patients and be extended to a more personalized therapeutic approach.
Keywords: chromosomal instability, acute myeloid leukemia, cytogenetic heterogeneity, aneuploidy, complex karyotype, TP53, centrosome dysfunction, MYC, telomere dysfunction, therapeutic targets, aging, synthetic lethality
1. Introduction
Since Boveri’s theory that chromosome abnormalities promote cancer, studies have attempted to elucidate the mechanisms behind the origins of chromosomal aberrations [1]. Chromosomal instability (CIN) is the increasing rate in which cells acquire new chromosomal alterations. Depending on the type of abnormalities, it can be classified into numerical CIN (nCIN), characterized by chromosome gains and losses, and structural CIN (sCIN) represented by chromosome translocations [2]. Importantly, CIN is one of the cancer hallmarks [3]. CIN can promote selective advantage to cancer cells by increasing the probability of novel chromosomal abnormalities, which can change the expression profile of the genes regulating cell division and differentiation, resulting in high proliferation rates [3,4]. Recent studies have shown a deep relationship of CIN with the origin, progression, and relapse in many cancers [5,6,7,8].
CIN not only occurs as a tumor-promotor mechanism but also as a tumor-suppressor mechanism. This observation comes from the evidence showing that different levels of CIN lead to distinct outcomes. Moderate or low levels of CIN are associated with increased rates of genetic cancer-promoting features. On the other hand, extreme levels of CIN could lead to decreased cell fitness or apoptosis [9]. The levels of CIN and the sites in which it occurs can also indicate different outcomes [10]. Therefore, CIN features not only could refine risk stratifications but also opens opportunities for new therapeutic approaches in cancer [9,11,12].
The current models for CIN involve telomere dysfunction, defective spindle assembly, sister chromatid cohesion, DNA double-strand breaks (DSB) repair, genes involved in the cell cycle, and epigenetic regulators. These CIN mechanisms and their signatures can be largely found in acute myeloid leukemia (AML), a heterogeneous disease characterized by abnormal proliferation and accumulation of myeloid precursor cells in the bone marrow [13]. AML can be classified as de novo AML, secondary AML (s-AML), whose origin is from a prior hematologic disease, and therapy-related AML (t-AML), which arises as a result of exposure to alkalizing agents, irradiation, and other factors associated to prior therapy [14,15]. Regardless of the classification, approximately 55% of AML patients show chromosomal abnormalities [16]. Cytogenetic abnormalities in AML are an important prognostic factor and are used for risk-stratification and guide treatment definition [17,18]. For example, a complex karyotype (CK) is associated with poor prognosis [19]. In older patients (≥60 years), only 10–44% of those with ≥3 cytogenetic abnormalities achieve complete remission (CR) after therapy, and for those with ≥5 chromosome abnormalities, the CR rates are significantly lower (7–26%) [20,21,22,23,24,25]. In this review, we will focus on the mechanisms associated with CIN resulting in cytogenetic abnormalities (Summarized in Figure 1), their prognostic impact, and the use of CIN as a target among the AML types.
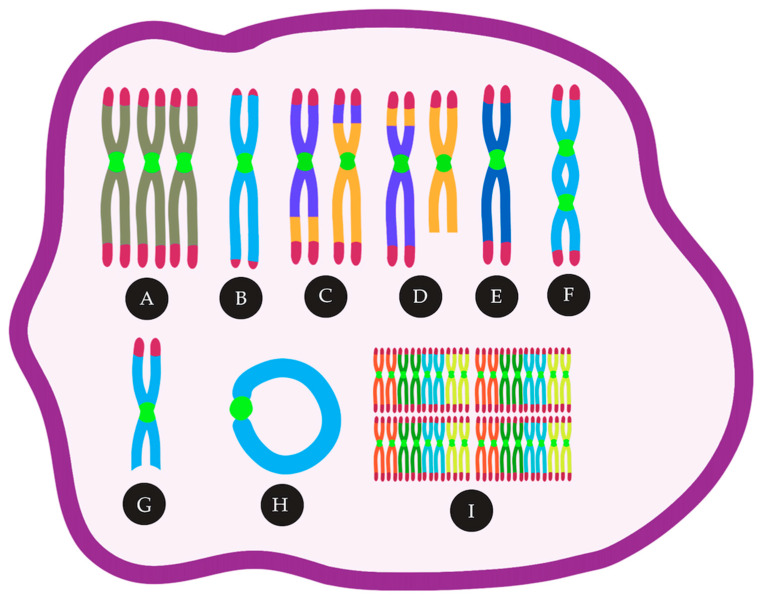
Figure 1.
CIN in AML can lead to many cytogenetic abnormalities, such as (A) trisomies, (B) telomere loss, (C) reciprocal translocations, (D) unbalanced translocations, (E) monosomies, (F) Dicentric chromosomes, (G) deletions, (H) ring chromosomes, (I) polyploidy.
2. Mechanisms and Consequences of CIN
2.1. Aneuploidy and CIN in AML, Mechanism and Consequence — An Inter-Relationship
One type of nCIN is aneuploidy, characterized by an abnormal number of chromosomes in the cell, which is a result of chromosome mis-segregation errors [26]. These errors can occur when the chromosomes fail to attach correctly to the mitotic spindle [27]. CIN and aneuploidy are not synonymous [28]. Although CIN is defined as the increasing rate in which cells acquire new chromosomal alterations, aneuploidy is associated to the abnormal number of chromosomes in the karyotype at a specific point in time [29]. Aneuploidy can also be found after clonal expansion, and this is not implying that the same cell will acquire new chromosomal alterations in every cell division. That is the case of constitutional trisomies, such as trisomy 21 or Down’s syndrome, which presents aneuploidy but not CIN [30]. However, patients with trisomy of the 21 have a predisposition to cancer, such as hematologic malignancies [31]. In AML, aneuploidy is present in more than 20% of cases [17,32,33,34] and is related to poor prognosis [17,33,35,36,37].
A key gene to maintain diploid karyotype in normal cells is TP53 [38]. This protein regulates cellular differentiation, cell cycle arrest, and DNA repair preventing genomic instability [39,40]. Mutations in TP53 arise before or after the first aneuploidy event [41,42] These mutations are also considered an early leukemogenic event in preleukemic stem cells [43,44]. Alteration in TP53 may result in the continued proliferation of aneuploidy cells or can trigger off apoptosis [45]. TP53 has a role in suppressing nCIN and sCIN by inducing apoptosis in cells that have a long pause in the mitotic checkpoint, which indicates DNA damage [46]. Mutations in tumor suppressor genes comprise 16% of AML patients, and TP53 is one of them [34]. The incidence of TP53 mutations in AML varies between 10% and 15% [34,47,48]. In general, TP53 mutations are highly present in AML patients with CK (60%) [47,49,50,51].
Interesting, the incidence of TP53 mutations in AML is low in comparison with other cancer types. This observation highlights that additional mechanisms are affecting p53 function for AML patients [52]. Many pathways have been proposed for non-mutational wtp53 inactivation in AML. Mdm4, a p53 negative regulator, is overexpressed in 10% of cases in wtp53 AML with CK [53,54]. Li et al. (2014) described that Mdm4 overexpression was associated with aneuploidy or polyploidy, showing an important link between Mdm4 overexpression, wtp53 inhibition and CIN in AML [53,54]. Additionally, Mdm2 overexpression, another p53 negative regulator, is seen in more than 50% of wtp53 AML patients [55,56,57]. Together with Mdm2 and Mdm4 overexpression, ARF down-regulation, deregulated post-translational modifications, and nuclear-cytoplasmic microRNAs were also described as non-mutational wtp53 inactivation in AML [52]. Cazzola et al. (2019) demonstrated that AML cells TP53-knockout have CIN phenotype and karyotype heterogeneity [58]. The deletion of 5q, a chromosomal region containing many protein-encoding genes associated with hematopoietic differentiation, together with TP53 mutations, can promote genome and chromosomal instability to block normal hematopoietic differentiation and is significantly associated with CK and poor prognostic [59,60,61]. In addition, 5q deletion, which leads to the haploinsufficiency of the genes involved in cell cycle control, as a sole chromosomal abnormality, is rare in AML patients. However, together with TP53, this alteration is frequently found in AML patients with CK (60–69%) [62,63,64]. Together, these observations show a strong association between CK and the deletion of checkpoint genes (TP53 and the ones located at 5q), suggesting an important role of dysfunctional cell cycle checkpoint in AML.
2.2. Chromosome Segregation Errors
2.2.1. Defects in the Spindle Assembly Checkpoint (SAC)
Jin and Burkard (2018) have associated CIN in AML patients with defects in the spindle assembly checkpoint (SAC). SAC is a mitotic checkpoint mechanism used to prevent transition to anaphase when there is an error on the kinetochore-microtubules attachments [65,66]. When SAC is malfunctioning, cells without proper spindle attachments can bypass anaphase checkpoints and divide [1]. The inactivation of the entire mitotic checkpoint can generate chromosome mis-segregation leading to CIN or cell death [67,68] in various cancers [69,70,71].
Mosaic variegated aneuploidy (MVA) is a rare human chromosomal instability disorder. Patients with MVA present germline mutations in the mitotic checkpoint components and 25% of the cells show aneuploidy [72]. The most relevant mutation is in the spindle checkpoint gene BUB1B, where the SAC protein BubR1 is expressed. BubR1 stabilizes the kinetochore-microtubule and corrects the proper chromosome positioning. It prevents cell division until forming an appropriate bi-oriented mitotic spindle [73]. In MVA, mutations in BUB1B leads to CIN with consecutive constitutional mosaicism for chromosomal gains and losses and subsequent predisposition to several types of cancer [74]. In AML, Schnerch et al. (2012) suggested a role of SAC insufficiency in the pathogenesis and progression of patients with a CK [75]. Since BUBR1 is an anaphase-promoting complex (APC/C) inhibitor gene, AML cells with defects in this gene usually allow chromosomal alterations to bypass mitosis [76].
The most common chromosomal abnormality found in AML is t(8;21)(q22;q22) [77]. Boyapati et al. (2007) demonstrated in cell lines that the resulting fusion protein t(8;21) from AML impairs the spindle checkpoint and promotes aneuploidy [78]. Nucleoporin 98 gene (NUP98) is another gene associated with SAC defects in AML. NUP98 regulates the timely destruction of securin by APC/C [79]. Cells with NUP98 translocation contain aberrant securin (a regulatory protein of the metaphase-anaphase transition), leading to aneuploidy [80]. Another APC/C protein with decreased expression in AML is Cdh1, an antagonist regulator of SAC (which activates and mediates securin degradation). Therefore, the high number of SAC alterations in AML cells can be associated with other dysfunctional chromosome segregation features.
2.2.2. Cohesion Defects
Sister chromatids are kept together until the proper formation of the bipolar spindle. If cohesion between sister chromatids is lost, chromosomes mis-segregate [81]. Cohesin defects, which is a protein complex mediating sister chromatid cohesion, are also associated with aneuploidy and CIN [82,83,84,85]. Ley et al. (2013), using whole-exome sequencing, showed mutations in cohesin complex genes in 13% of de novo AML patients [34]. However, cohesin gene mutations did not show a prognostic impact in AML. This is probably due to the co-existence of cohesin complex mutations with NPM1 (Nucleophosmin 1) mutations [86]. Interestingly, the co-existence of both mutations is associated with a favorable prognosis and normal karyotype in AML [87,88]. NPM1 mutation results in the inactivation of the nuclear factor-κB (NF-κB) in the cytoplasm [89]. Since activation of NF-κB provides drug resistance to chemotherapy drugs in AML, the association between cohesion and NPM1 mutations leads to favorable prognostic and chemosensitivity.
2.2.3. Centrosome Dysfunction and Assembly of Multipolar Mitotic Spindles
Another important mechanism related to CIN is centrosome dysfunction [90,91,92,93,94]. Dysfunctional centrosomes are characterized by the presence of aberrant centrosomes numbers, imbalances in centrosome-associated proteins expression, centrosome structural abnormalities, and alterations in the clustering of centrosomal components [95]. Cells presenting centrosome defects show the formation of multipolar mitotic spindles (cells with multiple centrosomes) [91,96]. Neben et al. (2003) have shown an association between abnormal centrosomes and the presence of cytogenetic alterations in AML. Interestingly, centrosome dysfunction allowed stratification into cytogenetic risk groups, where higher numbers of centrosome alterations were related to an increased adverse prognosis. The authors also suggested centrosome aberrations and multipolar mitotic spindles as the cause of numerical chromosome alterations in AML patients [97].
The aurora kinases, a family of serine-threonine protein kinases, have a key role in centrosome dynamics, mitotic spindle, and mitotic centrosomes [98]. Two common types of these proteins are Aurora A and Aurora B. Aurora A is active during the late S and early G2 phase and ensures a proper spindle assembly and chromosome alignment during mitosis [99]. On the other hand, Aurora B functions as a protein complex (through the G2 phase) and is in charge of bipolar attachment of the spindle to the centromeres and correct segregation of the sister chromatids [98]. Aurora kinases A and B are overexpressed in AML CD34+ blast cells compared to CD34+ from normal individuals with no evidence of hematologic diseases [100,101,102]. Lucena-Araujo et al.(2010) reported that high expression of Aurora Kinases A and B was related to unfavorable cytogenetic abnormalities, represented by CK and high blasts count in AML patients [103]. Yang et al. (2013) outlined that AML blasts overexpressing Aurora A were chemotherapy resistant. Aurora A negatively regulates p53 and, due to the role of p53 in the induction of apoptosis, its downregulation allows cells to escape from apoptosis induced by chemotherapy in AML [100].
2.3. DNA Double-strand Breaks
DSBs can lead to translocations and DNA deletions [104,105,106]. One of the major causes of DSBs is a failure in the chromosome decatenation (disentanglement of the chromosomes) [107,108,109]. In normal cells, the decatenation checkpoint during the G2 phase delays the entry into mitosis until every chromosome is decatenated by the enzyme topoisomerase IIα (topo II) [109,110]. Wray et al. (2009) reported that AML cells that fail to arrest at the mitotic decatenation checkpoint continue to proliferate due to Metnase activity. Metnase (SETMAR) is a SET-transposase fusion protein that promotes non-homologous end-joining repair even in the presence of Topo IIα inhibitor [111]. Jacoby et al. (2014) showed that DSBs response is abnormal in myeloblasts from t-AML patients, and this feature was associated with trisomy 8. The authors suggested that the association between abnormal DSBs and trisomy 8 in AML is related to MYC overexpression [112]. MYC is a proto-oncogene located at the long arm of chromosome 8 (8q24). MYC deregulation leads to DNA damage [113], the induction of genomic instability and telomere dysfunction [114]. The induction of DNA damage through DSBs seems to occur through direct MYC-mediated suppression of the NHEJ (Non-homologous end-joining), an important pathway that repairs DSBs in normal cells [115,116,117].
Trisomy 8 can be found in the blood of normal individuals [118,119]. Grove & Vassiliou (2014) proposed that it may be one of the early AML leukemogenesis events [120]. Importantly, the gain of chromosome 8 is one of the most common chromosomal abnormalities in AML. It represents 30–40% of cases alone or in association with other cytogenetic abnormalities, and it is known as the most frequent gain of chromosome in AML patients with CK [121,122,123].
2.4. Telomere Dysfunction
Telomeres are TTAGGG repetitive sequences directly associated with capping proteins, shelterin proteins, that protect the ends of chromosomes [124]. The linear chromosome DNA ends have a 3′ single-stranded overhang, which prevents those sites from being recognized as DSBs and activate DNA damage response pathways [125]. Telomere overhang length remains constant in healthy individuals over time [126,127,128]). However, in AML, Yan et al. (2013) reported that patients with abnormal karyotype presented shorter overhang length than those with normal karyotype [129]. The authors suggested the overhang length as an important prediction of poor prognosis in AML patients [129].
Telomeres become shorter at each cell division and, without telomerase, an enzyme that adds TTAGGG repetitive sequences to elongate the telomeres, cells undergo senescence [130]. The senescence occurs when telomeres become critically short, a phenomenon known as Hayflick limit, resulting in the cell cycle arrest. Nevertheless, cancer cells can bypass the telomere crisis through different mechanisms. Reactivation of telomerase is the most common mechanism to maintain telomere length, followed by the alternative mechanism of telomere lengthening (ALT) [131,132]. Telomerase reverse transcriptase (abbreviated as TERT, or hTERT in humans) is a catalytic subunit of the enzyme telomerase, which, together with the telomerase RNA component (TERC), comprises an important unit of the telomerase complex [133]. A decrease or loss of telomerase activity by mutations leads to telomere shortening, increasing the risk of CIN and, consequently, of cancer. The telomerase complex genes are frequently mutated in AML [134,135].
Swiggers et al. (2006) demonstrated that critically short telomeres in blasts AML patients lead to nCIN. The authors reported an increased rate of loss or gain of chromosome parts after telomere shortening [136]. Hartmann et al. (2005) also supported the relationship between short telomeres and CIN in AML. In both studies, telomere length and hTERT expression correlated with chromosomal abnormalities in AML patients. They found that telomere length in mononuclear cells of AML patients was significantly reduced compared to controls (peripheral blood granulocytes from healthy individuals). In addition, patients with abnormal karyotype presented shorter telomeres than those with normal karyotype. In contrast, extremely short telomeres (median of -3,7 kb compared to healthy donors) were found in AML patients showing multiple chromosomal abnormalities. Furthermore, hTERT continued to be associated with an increase in the karyotype complexity [137].
Capraro et al. (2012) have also shown that an abnormal karyotype was associated with shorter telomeres and extremely low telomerase activity in AML [138]. Interestingly, dyskeratosis congenita (DC), a disease characterized by bone marrow failure, also presents short telomeres and shows a high predisposition to AML (with approximately 200-fold for AML compared to the general population) [139]. Therefore, telomere shortening is viewed as an important feature in AML and to be related to poor prognosis [134,137,140,141]. However, Warny et al. (2019) reported data not corroborating with these previous studies. Their interesting findings point out that telomere length in the bone marrow mononuclear cells was similar in size both at the moment of diagnosis and at relapse. They also observed that telomere length increased after chemotherapy-induced remission, but no prognostic association was found [142]. Warny et al. (2019) correlated telomere maintenance with telomerase. This is an interesting observation since elevated telomerase activity and hTERT expression were reported in 87% of AML patients in another study [143]. Swiggers et al. (2006) also observed high telomerase activity, except that AML patients with short telomeres presented high telomerase activity. In this case, high expression of TRF1, a protein that is a negative regulator of telomere length, was proposed to explain the co-presence of high telomerase activity and extremely short telomeres [136,144].
Telomeres are also responsible for preventing end-to-end fusions of chromosomes, one of the major mechanisms that can trigger both nCIN and sCIN [145]. Critically short telomeres and, consequently, telomere aggregates can result in fused chromosomes with two centromeres (dicentric chromosomes) [146]. During anaphase, the dicentric chromosomes form a bridge between the bipolar spindles and the centromeres of the sister chromatids pulled in opposite directions causing their breakage. Importantly, such breaks can occur in different places of the chromosome, not necessarily between fused chromatids. The contiguous repetition of this process gives rise to the phenomenon known as breakage-fusion-bridge (BFB) cycles, which is associated with CIN [147,148].
Dicentric chromosomes (DC) are one of the major signatures of telomere dysfunction. The incidence of dicentric chromosomes varies among AML types, but in general is present in 8–15% of all AML cases [149,150]. DCs are mainly found in CK (23%), where more than one DC is usually present [59,151]. DCs play an important role in oncogenesis, as demonstrated by Gascoigne and Cheeseman (2013). The authors showed that the occurrence of a single dicentric chromosome could contribute to tumor initiation in AML [152]. Furthermore, Sarova et al. (2016) related the presence of DC to MDS progression to AML, in which the transformation was characterized by the acquisition of more complex karyotypes [151].
2.5. Complex Chromosomal Rearrangements
Complex chromosome rearrangements (CCR) have been extensively reported in AML [153,154,155,156]. Various mechanisms have been suggested to the occurrence of this phenomenon (e.g., non-homologous end joining (NHEJ), replication-based mechanisms, BBF cycles, telomere dysfunction) [157]. Some authors have been using the term chromothripsis for the event where genetic material suffers an enormous clustered chromosomal rearrangement on specific regions of one or few chromosomes in a single cell cycle [158]. Since chromothripsis is not proven to be the cause of this phenomenon, here we will describe these abnormalities just as CCRs [157]. Rausch et al. (2012) showed that in their cohort of AML TP53 mutated patients, ∼47% of cases presented CCRs. The occurrence of CCRs was associated with a poor prognosis [153]. Rücker et al. (2018) reported CCRs in 35% of AML patients with CK. In 85% of cases with CCRs presented mutated TP53 [159]. Once more, this data highlights the role of dysfunctional TP53 on CIN in AML. Hence, Fontana et al. (2018) have found an incidence of 6.6% CCRs in a large cohort of AML patients (N=395). It was also reported that AML cells with CCRs also presented signatures of CIN, such as TP53 alteration, a higher mean of copy number alteration (CNA), CK, 5q deletion, alterations in DNA repair, and cell cycle. They also observed that AML cells with CCRs had marker chromosomes with the MYC gene [155]. Furthermore, Gao et al. (2020) reported that in AML-MRC with CCRs, this phenomenon was associated with a lower number of white blood cells and platelets and a higher degree of karyotypic complexity. The most involved chromosomes in CCRs were the chromosomes 8 and 11, resulting in the amplification of MYC (8q24.2) or lysine methyltransferase 2A (KMT2A) (11q23.3) [160]. L′Abbate et al. (2018) analyzed MYC amplicons in AML. Their results provide evidence that CCRs are not related to a single catastrophic event as the chromothripsis model describes it but rather to an accumulative evolution [161]. Marker chromosomes are rearranged chromosomes whose genetic origin cannot be verified by conventional banding cytogenetics techniques [162]. In AML, Bochtler et al. (2017) reported that marker chromosomes could arise from CCRs and predict adverse prognosis [154]. Marker chromosomes were also suggested to be a risk classification factor for AML with adverse cytogenetics [163].
2.6. Epigenetic Regulation
Abnormalities in the epigenetic regulator Tet methylcytosine dioxygenase 2 (TET2) and Enhancer of zeste homolog 2 (EZH2) could induce CIN through the deregulation of histone modifications, which alters the chromatin structure and affect gene expression [164,165,166]. Mutations in the TET2 are among the most common mutations in AML [167,168,169]. EZH2 is located in 7q36.1, a chromosomal region affected by the loss of chromosome 7 (-7) or deletion of 7q, which reduces its gene expression [170,171]. The -7 and deletion of 7q are highly associated with CK and adverse prognosis [50,172]. Wang et al. (2020) reported that TET2 is hypermethylated (downregulated transcription) in 30% of AML patients. Alterations in the expression of TET2 or EZH2 also protects against apoptosis by an unknown mechanism [173]. Göllner et al. (2017) showed that EZH2 loss of function induced resistance to multiple drugs in AML [174]. Interestingly, the expression levels of the genes TET2 and EZH2 were also positively correlated to the CIN MAD2 and CDC20 genes expression levels [173]. The protein mitotic arrest deficient 2 (Mad2) and cell division cycle protein 20 homologue (CDC20) overexpression and downregulation are frequently altered in many cancers and associated with CIN. Both proteins are essential for the mitotic checkpoint, in which they act together as an APC/C inhibitor, preventing aneuploidy and, consequently, CIN [1,175,176]. Schvartzman et al. (2011) have shown, in a p53 mutant tumor model, that wtp53 represses mad2 and its upregulation is necessary for CIN in AML [174]. Overexpression of CDC20 is more present in aneuploid than euploid AML [177].
3. Clinical Considerations
A summary of CIN features and the studies associating them to AML pathogenesis, evolution to worst outcomes, and relapse are shown in Table 1. It is important to mention that the incidence of AML increases with age (mean age of 75 years) [13]. An example of how age affects AML prognosis is observed in acute promyelocytic leukemia (APL), a subtype of AML characterized by the presence of t(15;17). Patients with APL present median age of 41–48 years, having a favorable prognosis and rare CK incidence (only 4% of the cases) [178,179]. CIN increases with age due to accumulation of genetic abnormalities, defective DNA damage response, and bone marrow loss of function (decreased repopulate ability of the hematopoietic system) [180,181]. Indeed CK, aneuploidy, telomere dysfunction, abnormal epigenetics, MYC, and TP53 abnormalities are all age-related in AML [20,21,25,182,183,184]. The prognosis of sAML and tAML, in general, is poor when compared to de novo AML. However, the de novo AML patients older than 60 years of age and adverse cytogenetics present a similar prognosis of sAML and tAML [185]. The three AML types also shows differences in the number of CIN features found (Table 2).
Table 1.
CIN features during AML course and drugs used in preclinical and clinical studies.
| Features | Promising Agents 1 | |
|---|---|---|
| CK | [20,186,187] | PLK1 inhibitors [188]. In TP53 mutated: APR-246 [189] |
| Aneuploidy | [190,191] | PLK1 inhibitors [188]. In TP53 mutated: APR-246 [189] |
| TP53 | [192,193,194] | APR-246 with azacitidine (AZA) [195,196]. APR-246 alone [189], APR-246 combined with azacitidine [197,198]. |
| Abnormal 5q | [64,187,199,200] | PLK1 inhibitors [188]. In TP53 mutated: APR-246 [189] APR-246 with azacitidine [195,196]. APR-246 alone [189] |
| MYC abnormalities | [201,202] | Molibresib (GSK525762) [203,204] JQ1 combined with All-trans retinoic acid (ATRA) [205] Dihydroergotamine (DHE) [206] |
| Trisomy 8 | [118,119,120,121] | Molibresib (GSK525762) [203,204] JQ1 combined with All-trans retinoic acid (ATRA) [205] Dihydroergotamine (DHE) [206] |
| Telomere dysfunction | [136,144] | (hTERT) JQ1 combined with All-trans retinoic acid (ATRA) [205] Telomerase inhibitors [201,207]. |
| Dicentric chromosomes (DCs) | [150,208] | PLK1 inhibitors [188]. In TP53 mutated: APR-246 [189] APR-246 with azacitidine [195,196]. APR-246 alone [189] |
| Aurora Kinases | [100,101,102,103,209] | Small molecule inhibitors [210,211,212,213,214,215,216,217,218,219,220] |
| TET2 abnormalities | [173,221,222] | Decitabine [173]. Vitamin C combined with PARP inhibitor [223]. Ascorbate (Vitamin C) [224] |
| EZH2 abnormalities | [173,225] | Decitabine [173] |
1. Preclinical and clinical studies.
Table 2.
Incidence of CIN features in different types of AML.
| Features | de novo AML | sAML | tAML | References |
|---|---|---|---|---|
| Chromosomal abnormalities | 47–51% | 62.2% | 75–92% | [185,226,227] |
| Adverse cytogenetics | 19% | 22% | 39–40% | [185,226] |
| Complex karyotype (CK) | 10–23% | 18% | 26- 40% | [24,25,226] |
| Abnormal 17p | 5–11% | 14% | 14% | [33,226,228] |
| TP53 Mutation | 7–21% | 15% | 47% | [192,229] |
| −5 or 5q− | 5–8% | 7–14% | 14–21% | [25,33,226,227] |
| Abnormal chromosome 7 | 2–7% | 19% | 10% | [25,33,226,230] |
| Telomere Length: TRF | No significant differences | ND | [141,231] | |
| MYC overexpression | 35% | 27% | ND | [232] |
| +8 | 10–25% | 25% | 26% | [228,233] |
| Dicentric Chromosomes (DCs) | 3% | ND | 15% | [149] |
| DCs in CK cohorts | 48% | 59% | ND | [208] |
| TET2 mutations | 13–27% | 24–29% | 24% | [222,234,235] |
| EZH2 abnormalities | 2–4% | 7–9% | 8% | [229,236,237] |
ND: not described.
Beyond assessing CIN in vitro in patient tumor samples to predict prognosis, current and emerging AML therapies are exploring CIN to improve AML treatment. Small-molecule inhibitors of aurora kinases are in phase I/II clinical trials for AML [210,211,213,214,215,216,217,218,219]. Yang et al. (2013) demonstrated that the proliferation of AML cells overexpressing Aurora A was significantly decreased by using aurora inhibitors such as alisertib (MLN8273), an oral aurora A kinase inhibitor. The authors noticed that the reduced proliferation was linked to a decrease in the self-renewal capability of AML blasts and apoptosis induction [100]. Besides, Kelly et al. (2012) reported that alisertib also enhanced cytarabine’s efficacy, a frontline chemotherapy medication used to treat AML. Cytarabine inhibits the DNA synthesis and, together with alisertib, generates intense stress response, which explains the better efficacy of the combination [238]. Recently, Brunner et al. (2018), in a phase II trial, showed that 67% of AML patients with adverse karyotype and 75% of those with TP53 mutations achieved complete remission with the use of alisertib [207].
Moreover, Ikezoe et al. (2010) have shown that the induction of apoptosis by barasertib (AZD1152), an aurora B inhibitor, is dependent on functional p53. One of the consequences of using barasertib is the generation of polyploid cells. Aurora B inhibition leads to cells directly re-entering the S-phase without the cytokinesis. Barasertib aurora B inhibition in wild-type p53 cancer cells leads to increased p53 protein level and expression of p53 target genes to inhibit tumor growth [220,239]. Taken together, these results suggest that aurora kinases are promising targets for the elimination of chemotherapy-resistant AML blast cells since alterations in TP53 are present in only 10–15% of AML patients. Interestingly, in acute megakaryocytic leukemia (AMKL), characterized by the presence of abnormal megakaryoblasts, aurora inhibitors can induce blasts differentiation due to their properties of inducing polyploidy (normal megakaryocytes are polyploidy cells). Aurora inhibitors in AMKL induce the upregulation of CD41 and CD42 expression, two characteristic markers of differentiated megakaryocytes [240,241].
With respect to telomere dysfunction, AML cells could be targeted by telomerase inhibitors [201,207]. Bruedigam et al. (2014) demonstrated that in xenografts of primary human AML, the pharmacological or genetic inhibition of telomerase targets AML cells, decreases leukemia progression, and detains relapse following chemotherapy [242]. The authors used Imetelstat, a potent and specific telomerase inhibitor under clinical trials in many cancers [243]. Rusbuldt et al. (2017) also showed promising results of Imetelstat in AML. Combination of this drug with Venetoclax, an approved BCL-2 inhibitor used to treat chronic lymphocytic leukemia, showed a synergistic effect on apoptosis both in cell lines and patient samples in vitro, with a prolonged survival in xenograft models [244]. DiNardo et al. (2020) in a phase 3 trial, demonstrated that Venetoclax combined with azacitidine improved overall survival and remission when compared to those treated only with azacytidine [212]. MYC overexpression can also be a target, as was demonstrated in preclinical studies using molibresib (GSK525762), an orally bioavailable drug, which reduced c-MYC expression and its downstream transcriptional effects [245]. This drug is currently under investigation to treat AML patients [204]. MYC inhibition could overcome resistance to cytotoxic drugs in AML cells by promoting differentiation [202]. Another small molecule inhibitor that targets MYC is JQ1 [203]. JQ1 combined with all-trans retinoic acid (ATRA) synergistically suppresses AML cells proliferation [205]. The combination of ATRA and JQ1, which targets MYC and hTERT, could be useful especially for patients with telomere dysfunction [205].
Moreover, TET2 function can be restored by using ascorbate, a potentially non-toxic therapy that promotes DNA demethylation, differentiation, and cell death in leukemic cells [223,246,247,248]. This approach enhances leukemic cells sensitivity to PARP inhibitors [223]. The use of ascorbate has been associated with better outcomes for AML patients with TET2 mutations [224,249].
Regarding patients with mutations in TP53, a phase II study performed by Cluzeau et al. (2019) demonstrated that the combination of APR-246 (a drug that reactivates the mutated p53) with azacitidine (a DNA methyltransferase inhibitor whose cytotoxicity interferes with DNA synthesis) showed a response rate of 75% including 56% of complete remission (CR) [195,196]. Polo-like kinase 1(PLK1), one important mitotic regulator overexpressed in AML, could also be targetable [250,251,252]. Moison et al. (2012) have shown that PLK1 inhibitors induce apoptosis in mutated and wild-type TP53 cells with complex karyotype in AML [188]. PLK1 inhibitors are in clinical studies for AML [253].
Targeting CIN as an approach to AML therapy is another attractive strategy, but some drugs could increase CIN beyond the “accepted” threshold and actually induce cell death. Jin & Burkard (2018) [65] measured CIN by verifying the chromosome mis-segregation frequency in AML samples and demonstrated that high levels of CIN were correlated with better overall survival. They also induced CIN with AZ3146 (an inhibitor of the Mps1 mitotic checkpoint kinase) in AML cell lines and showed that Mps1 inhibition induced a robust type I interferon (I IFN) response. This response is known to trigger the activation of many immune system cells, promote exposition of antigen, and enhance T-cell responses. Therefore, high CIN and Mps1 inhibitors seem to be promising therapies in AML [19]. The lack of I IFN response is known to cause resistance against chemotherapy in other cancers, which is overcome by supplying I IFN [254]. The use of type I interferon to treat AML is also suggested by other studies [255,256,257]. Furthermore, the role of high levels of CIN as a cancer suppressor has been reported on gastric, non-small cell lung, ovarian, and ER-negative breast cancers [258,259,260].
High degrees of CIN can also induce cell death through a mechanism known as synthetic lethality. Synthetic lethality is characterized by simultaneous mutations in two different genes, resulting in a significant decrease in cell fitness compared with the same mutations occurring independently [11,261]. Synthetic lethal approaches are already being applied to AML studies and show promising results. One of the main causes of drug resistance in AML is apoptosis evasion [262]. Pan et al. (2017), using AML resistant mouse models, demonstrated that synthetic lethality induced by the combination of Bcl-2 inhibition (an anti-apoptotic protein) and p53 activation overcome apoptosis resistance in AML [263]. Leukemic cells with mutated BRCA1 are more dependent on functional poly (ADP-ribose) polymerase (PARP) proteins than normal cells. They depend on PARP’s DNA repair role by homologous recombination (HR) [264]. PARP inhibitors can be used to selectively kill BRCA1- or BRCA2-mutated cells in AML BRCA1 mutated patients (BRCA1 loss is reported in 12% of AML patients) [168]. Furthermore, defective HR also provides the potential use of PARP inhibitors targeting other key components of HR [265]. In AML, the AML1-ETO fusion, a product of the chromosomal translocation t(8;21) (q22;q22), results in the repression of genes that are essential to the DNA damage response [266,267]. Esposito et al. (2015) demonstrated, both in vitro and in vivo, that the suppression of HR transcriptional programs in AML1-ETO or PML-RARa cells leads to sensitivity to PARP inhibitors [268]. Faraoni et al. (2015) has shown in vitro that the use of the PARP inhibitor Olaparib induced cell death in cell lines with 11q23 deletion [269]. They also demonstrated that this drug selectively killed leukemic blasts, not affecting normal bone marrow CD34 cells [269].
4. Conclusions
The clinical outcome of AML is very heterogeneous and can range from few days of median survival to complete cure, depending on AML subgroups. In AML, the karyotype is the most powerful predictor of treatment outcome. Approximately 30% of cases of AML have an unfavorable karyotype and, if treated with conventional chemotherapy, a 5-year overall survival of 10% to 20% is expected. The best chance of cure for those patients seems to be an allogeneic transplant, but elderly patients are not eligible. Several studies had highlighted CIN as an important mechanism related to the origin, evolution, and relapse of AML. AML patients with CK can present various CIN features, such as defects in the spindle assembly checkpoint, centrosome dysfunction and assembly of multipolar mitotic spindles, defective DNA damage response, telomere dysfunction, and chromosomal abnormalities known to trigger CIN (Figure 2). CIN signatures can occur in all the AML ontogenies, but their incidence varies among de novo AML, sAML, and tAML. The higher incidence of chromosomal abnormalities found in tAML can be correlated to the prevalence of CIN features in this AML entity and results in the poor prognostic among the AML types. The CIN features in AML are strongly related to ageing, prior disease characterized by CIN, and previous exposure to cytotoxic therapy. Although most of these features show poor prognostic value, they also offer strong new perspectives on personalized therapeutic approaches to decrease the toxic effects of chemotherapy and relapse rates in AML patients. For each CIN feature occurring in AML, there are studies in different research phases showing promising results on targeting those specific mechanisms. Therefore, these studies open the perspective of combining multiple therapies based on the occurrence of the different CIN features individually or on their co-occurrence. The characterization of CIN features that are not in the current routine analyses of AML patients might provide a better stratification of patients. Studies based on the CIN approach with a more extensive characterization of CIN signatures may provide better therapy strategies, especially for those patients with a high risk of relapse or who do not respond properly to current chemotherapy. Future studies evaluating the association among the mechanisms cited here comprise venues for further exploration.
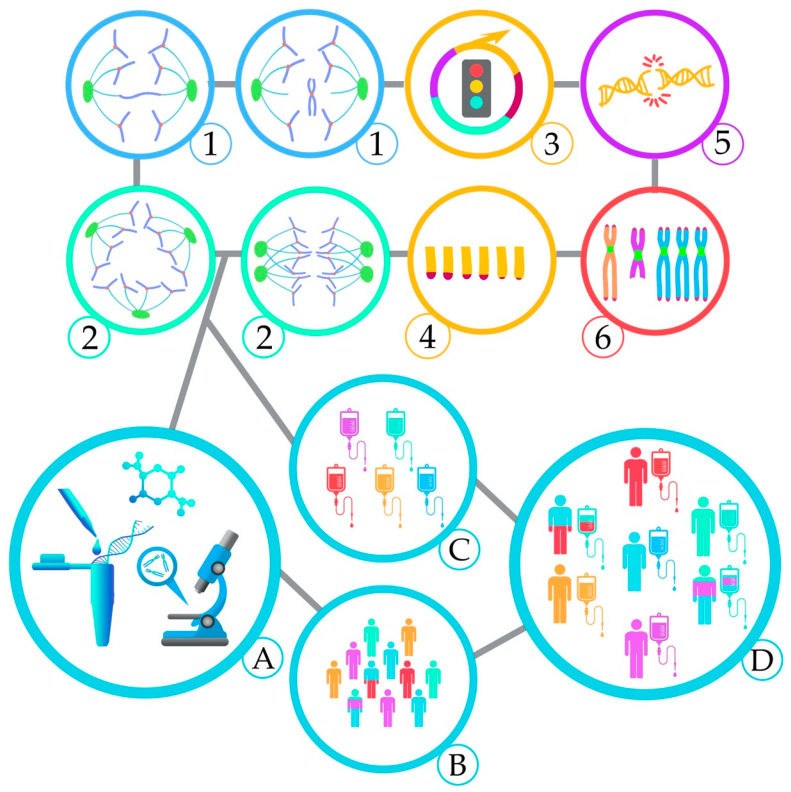
Figure 2.
AML patients can present various CIN features, such as Chromosome mis-segregation (1); Multipolar spindles (2); Dysfunctional cell cycle checkpoints (3); Telomere dysfunction (4); Defective DNA damage response (5); Chromosomal abnormalities associated with CIN (6). These unique CIN markers could be used in clinical practice (A) for better stratification of patients with complex karyotype (B) and combine new target therapies (C) for personalized treatments (D).
Acknowledgments
The authors would like to thank Elizabete Cruz for helping in the manuscript preparation.
Author Contributions
Writing—original draft preparation, M.d.O.L.; writing—review and editing, P.R.S.B.; A.T.S.-B.; A.R.-P., and S.M.; visualization, M.d.O.L. and A.R.-P.; supervision, P.R.S.B.; A.R.-P., and S.M.; funding acquisition, S.M. All authors have read and agreed to the published version of the manuscript.
Funding
The authors thank the Canadian Institutes of Health Research (CIHR) for CRC funding (S.M.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holland A.J., Cleveland D.W. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakhoum S.F., Kabeche L., Murnane J.P., Zaki B.I., Compton D.A. DNA-Damage Response during Mitosis Induces Whole-Chromosome Missegregation. Cancer Discov. 2014;4:1281–1289. doi: 10.1158/2159-8290.CD-14-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Targa A., Rancati G. Cancer: A CINful evolution. Curr. Opin. Cell Biol. 2018;52:136–144. doi: 10.1016/j.ceb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Cahill D.P., Kinzler K.W., Vogelstein B., Lengauer C. Genetic instability and darwinian selection in tumours. Trends Genet. 1999;15:M57–M60. doi: 10.1016/S0168-9525(99)01874-0. [DOI] [PubMed] [Google Scholar]
- 5.Bakhoum S.F., Cantley L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A.J.X., Endesfelder D., Rowan A.J., Walther A., Birkbak N.J., Futreal P.A., Downward J., Szallasi Z., Tomlinson I.P.M., Howell M., et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bach D.-H., Zhang W., Sood A.K. Chromosomal Instability in Tumor Initiation and Development. Cancer Res. 2019;79:3995–4002. doi: 10.1158/0008-5472.CAN-18-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salgueiro L., Buccitelli C., Rowald K., Somogyi K., Kandala S., Korbel J.O., Sotillo R. Acquisition of chromosome instability is a mechanism to evade oncogene addiction. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.201910941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silk A.D., Zasadil L.M., Holland A.J., Vitre B., Cleveland D.W., Weaver B.A. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. USA. 2013;110:E4134–E4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoevenaar W.H.M., Janssen A., Quirindongo A.I., Ma H., Klaasen S.J., Teixeira A., van Gerwen B., Lansu N., Morsink F.H.M., Offerhaus G.J.A., et al. Degree and site of chromosomal instability define its oncogenic potential. Nat. Commun. 2020;11:1501. doi: 10.1038/s41467-020-15279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy S., Fam H.K., Wang Y.K., Styles E.B., Kim J.-H., Ang J.S., Singh T., Larionov V., Shah S.P., Andrews B., et al. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc. Natl. Acad. Sci. USA. 2016;113:9967–9976. doi: 10.1073/pnas.1611839113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vargas-Rondón N., Villegas V., Rondón-Lagos M. The Role of Chromosomal Instability in Cancer and Therapeutic Responses. Cancers. 2017;10:4. doi: 10.3390/cancers10010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estey E., Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 14.Hulegårdh E., Nilsson C., Lazarevic V., Garelius H., Antunovic P., Rangert Derolf Å., Möllgård L., Uggla B., Wennström L., Wahlin A., et al. Characterization and prognostic features of secondary acute myeloid leukemia in a population-based setting: A report from the Swedish Acute Leukemia Registry. Am. J. Hematol. 2015;90:208–214. doi: 10.1002/ajh.23908. [DOI] [PubMed] [Google Scholar]
- 15.Sill H., Olipitz W., Zebisch A., Schulz E., Wölfler A. Therapy-related myeloid neoplasms: Pathobiology and clinical characteristics. Br. J. Pharmacol. 2011;162:792–805. doi: 10.1111/j.1476-5381.2010.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mrózek K., Heerema N.A., Bloomfield C.D. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 17.Grimwade D., Mrózek K. Diagnostic and Prognostic Value of Cytogenetics in Acute Myeloid Leukemia. Hematol. Oncol. Clin. 2011;25:1135–1161. doi: 10.1016/j.hoc.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin N., Lera R.F., Yan R.E., Guo F., Oxendine K., Horner V.L., Hu Y., Wan J., Mattison R.J., Weaver B.A., et al. Chromosomal instability upregulates interferon in acute myeloid leukemia. Genes Chromosom. Cancer. 2020:gcc.22880. doi: 10.1002/gcc.22880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mrózek K. Cytogenetic, Molecular Genetic, and Clinical Characteristics of Acute Myeloid Leukemia with a Complex Karyotype. Semin. Oncol. 2008;35:365–377. doi: 10.1053/j.seminoncol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fröhling S., Schlenk R.F., Kayser S., Morhardt M., Benner A., Döhner K., Döhner H. Cytogenetics and age are major determinants of outcome in intensively treated acute myeloid leukemia patients older than 60 years: Results from AMLSG trial AML HD98-B. Blood. 2006;108:3280–3288. doi: 10.1182/blood-2006-04-014324. [DOI] [PubMed] [Google Scholar]
- 22.Farag S.S., Archer K.J., Mrózek K., Ruppert A.S., Carroll A.J., Vardiman J.W., Pettenati M.J., Baer M.R., Qumsiyeh M.B., Koduru P.R., et al. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: Results from Cancer and Leukemia Group B 8461. Blood. 2006;108:63–73. doi: 10.1182/blood-2005-11-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Der Holt B., Breems D.A., Berna Beverloo H., Van Den Berg E., Burnett A.K., Sonneveld P., Löwenberg B. Various distinctive cytogenetic abnormalities in patients with acute myeloid leukaemia aged 60 years and older express adverse prognostic value: Results from a prospective clinical trial. Br. J. Haematol. 2007;136:96–105. doi: 10.1111/j.1365-2141.2006.06403.x. [DOI] [PubMed] [Google Scholar]
- 24.Schoch C., Haferlach T., Haase D., Fonatsch C., Loffler H., Schlegelberger B., Staib P., Sauerland M.C., Heinecke A., Buchner T., et al. Patients with de novo acute myeloid leukaemia and complex karyotype aberrations show a poor prognosis despite intensive treatment: A study of 90 patients. Br. J. Haematol. 2001;112:118–126. doi: 10.1046/j.1365-2141.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 25.Grimwade D. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): Analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.V98.5.1312. [DOI] [PubMed] [Google Scholar]
- 26.Wei W., Cheng Y., Wang B. Genome Stability. Elsevier; Amsterdam, The Netherlands: 2016. Cancer and Genomic Instability; pp. 463–486. [Google Scholar]
- 27.Compton D.A. Mechanisms of aneuploidy. Curr. Opin. Cell Biol. 2011;23:109–113. doi: 10.1016/j.ceb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schukken K.M., Foijer F. CIN and Aneuploidy: Different Concepts, Different Consequences. BioEssays. 2018;40:1700147. doi: 10.1002/bies.201700147. [DOI] [PubMed] [Google Scholar]
- 29.Geigl J.B., Obenauf A.C., Schwarzbraun T., Speicher M.R. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Hitzler J.K., Zipursky A. Origins of leukaemia in children with Down syndrome. Nat. Rev. Cancer. 2005;5:11–20. doi: 10.1038/nrc1525. [DOI] [PubMed] [Google Scholar]
- 31.Ganmore I., Smooha G., Izraeli S. Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 2009;18:R84–R93. doi: 10.1093/hmg/ddp084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breems D.A., Van Putten W.L.J., De Greef G.E., Van Zelderen-Bhola S.L., Gerssen-Schoorl K.B.J., Mellink C.H.M., Nieuwint A., Jotterand M., Hagemeijer A., Beverloo H.B., et al. Monosomal Karyotype in Acute Myeloid Leukemia: A Better Indicator of Poor Prognosis Than a Complex Karyotype. J. Clin. Oncol. 2008;26:4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
- 33.Grimwade D., Hills R.K., Moorman A.V., Walker H., Chatters S., Goldstone A.H., Wheatley K., Harrison C.J., Burnett A.K. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 34.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson G., Hoadley K., Triche T.J., Laird P.W., Baty J.D., et al. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farag S., Archer K., Mrozek K., Vardiman J., Carroll A., Pettenati M., Moore J., Kolitz J., Mayer R., Stone R., et al. Isolated trisomy of chromosomes 8, 11, 13 and 21 is an adverse prognostic factor in adults with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B 8461. Int. J. Oncol. 2002;21:1041–1051. doi: 10.3892/ijo.21.5.1041. [DOI] [PubMed] [Google Scholar]
- 36.Herold T., Metzeler K.H., Vosberg S., Hartmann L., Röllig C., Stölzel F., Schneider S., Hubmann M., Zellmeier E., Ksienzyk B., et al. Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood. 2014;124:1304–1311. doi: 10.1182/blood-2013-12-540716. [DOI] [PubMed] [Google Scholar]
- 37.Perrot A., Luquet I., Pigneux A., Mugneret F., Delaunay J., Harousseau J.-L., Barin C., Cahn J.-Y., Guardiola P., Himberlin C., et al. Dismal prognostic value of monosomal karyotype in elderly patients with acute myeloid leukemia: A GOELAMS study of 186 patients with unfavorable cytogenetic abnormalities. Blood. 2011;118:679–685. doi: 10.1182/blood-2010-09-307264. [DOI] [PubMed] [Google Scholar]
- 38.Thompson S.L., Compton D.A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahl G.M., Linke S.P., Paulson T.G., Huang L.C. Maintaining genetic stability through TP53 mediated checkpoint control. Cancer Surv. 1997;29:183–219. [PubMed] [Google Scholar]
- 40.Vousden K.H., Lu X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 41.Blount P.L., Galipeau P.C., Sanchez C.A., Neshat K., Levine D.S., Yin J., Suzuki H., Abraham J.M., Meltzer S.J., Reid B.J. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–2295. [PubMed] [Google Scholar]
- 42.Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 43.Reinisch A., Chan S.M., Thomas D., Majeti R. Biology and Clinical Relevance of Acute Myeloid Leukemia Stem Cells. Semin. Hematol. 2015;52:150–164. doi: 10.1053/j.seminhematol.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal R., Lind K., Heitzer E., Ulz P., Aubell K., Kashofer K., Middeke J.M., Thiede C., Schulz E., Rosenberger A., et al. Somatic TP53 mutations characterize preleukemic stem cells in acute myeloid leukemia. Blood. 2017;129:2587–2591. doi: 10.1182/blood-2016-11-751008. [DOI] [PubMed] [Google Scholar]
- 45.Fukasawa K., Wiener F., Woude G.F.V., Mai S. Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene. 1997;15:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 46.Dalton W.B., Yu B., Yang V.W. p53 suppresses structural chromosome instability after mitotic arrest in human cells. Oncogene. 2010;29:1929–1940. doi: 10.1038/onc.2009.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stengel A., Kern W., Haferlach T., Meggendorfer M., Fasan A., Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: An analysis of 3307 cases. Leukemia. 2017;31:705–711. doi: 10.1038/leu.2016.263. [DOI] [PubMed] [Google Scholar]
- 48.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haferlach C., Dicker F., Herholz H., Schnittger S., Kern W., Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22:1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 50.Rücker F.G., Schlenk R.F., Bullinger L., Kayser S., Teleanu V., Kett H., Habdank M., Kugler C.-M., Holzmann K., Gaidzik V.I., et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 51.Bowen D., Groves M.J., Burnett A.K., Patel Y., Allen C., Green C., Gale R.E., Hills R., Linch D.C. TP53 gene mutation is frequent in patients with acute myeloid leukemia and complex karyotype, and is associated with very poor prognosis. Leukemia. 2009;23:203–206. doi: 10.1038/leu.2008.173. [DOI] [PubMed] [Google Scholar]
- 52.Prokocimer M., Molchadsky A., Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood. 2017;130:699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan B.X., Khoo K.H., Lim T.M., Lane D.P. High Mdm4 levels suppress p53 activity and enhance its half-life in acute myeloid leukaemia. Oncotarget. 2014;5:933–943. doi: 10.18632/oncotarget.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Tan Y., Chen X., Xu Z., Yang S., Ren F., Guo H., Wang X., Chen Y., Li G., et al. MDM4 Overexpressed in Acute Myeloid Leukemia Patients with Complex Karyotype and Wild-Type TP53. PLoS ONE. 2014;9:e113088. doi: 10.1371/journal.pone.0113088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bueso-Ramos C., Yang Y., DeLeon E., McCown P., Stass S., Albitar M. The human MDM-2 oncogene is overexpressed in leukemias. Blood. 1993;82:2617–2623. doi: 10.1182/blood.V82.9.2617.2617. [DOI] [PubMed] [Google Scholar]
- 56.Faderl S., Kantarjian H.M., Estey E., Manshouri T., Chan C.-Y., Rahman Elsaied A., Kornblau S.M., Cortes J., Thomas D.A., Pierce S., et al. The prognostic significance of p16 INK4a /p14 ARF locus deletion and MDM-2 protein expression in adult acute myelogenous leukemia. Cancer. 2000;89:1976–1982. doi: 10.1002/1097-0142(20001101)89:9<1976::AID-CNCR14>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 57.Quintás-Cardama A., Hu C., Qutub A., Qiu Y.H., Zhang X., Post S.M., Zhang N., Coombes K., Kornblau S.M. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2017;31:1296–1305. doi: 10.1038/leu.2016.350. [DOI] [PubMed] [Google Scholar]
- 58.Cazzola A., Schlegel C., Jansen I., Bochtler T., Jauch A., Krämer A. TP53 deficiency permits chromosome abnormalities and karyotype heterogeneity in acute myeloid leukemia. Leukemia. 2019;33:2619–2627. doi: 10.1038/s41375-019-0550-5. [DOI] [PubMed] [Google Scholar]
- 59.Wang P., Spielberger R.T., Thangavelu M., Zhao N., Davis E.M., Iannantuoni K., Larson R.A., Le Beau M.M. dic(5;17): A recurring abnormality in malignant myeloid disorders associated with mutations ofTP53. Genes Chromosom. Cancer. 1997;20:282–291. doi: 10.1002/(SICI)1098-2264(199711)20:3<282::AID-GCC9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 60.Christiansen D.H., Andersen M.K., Pedersen-Bjergaard J. Mutations with Loss of Heterozygosity of p53 Are Common in Therapy-Related Myelodysplasia and Acute Myeloid Leukemia After Exposure to Alkylating Agents and Significantly Associated With Deletion or Loss of 5q, a Complex Karyotype, and a Poor Prognosis. J. Clin. Oncol. 2001;19:1405–1413. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- 61.Reilly A., Busch S., Abkowitz J.L., Becker P.S., Doulatov S. Deletions of 5q Promote Genome Instability in TP53-Mutant MDS/AML. Blood. 2019;134:1244. doi: 10.1182/blood-2019-127038. [DOI] [Google Scholar]
- 62.Schoch C., Haferlach T., Bursch S., Gerstner D., Schnittger S., Dugas M., Kern W., Löffler H., Hiddemann W. Loss of genetic material is more common than gain in acute myeloid leukemia with complex aberrant karyotype: A detailed analysis of 125 cases using conventional chromosome analysis and fluorescence in situ hybridization including 24-color FISH. Genes Chromosom. Cancer. 2002;35:20–29. doi: 10.1002/gcc.10088. [DOI] [PubMed] [Google Scholar]
- 63.Sanderson R.N., Johnson P.R.E., Moorman A.V., Roman E., Willett E., Taylor P.R., Proctor S.J., Bown N., Ogston S., Bowen D.T. Population-based demographic study of karyotypes in 1709 patients with adult Acute Myeloid Leukemia. Leukemia. 2006;20:444–450. doi: 10.1038/sj.leu.2404055. [DOI] [PubMed] [Google Scholar]
- 64.Volkert S., Kohlmann A., Schnittger S., Kern W., Haferlach T., Haferlach C. Association of the type of 5q loss with complex karyotype, clonal evolution, TP53 mutation status, and prognosis in acute myeloid leukemia and myelodysplastic syndrome. Genes Chromosom. Cancer. 2014;53:402–410. doi: 10.1002/gcc.22151. [DOI] [PubMed] [Google Scholar]
- 65.Jin N., Burkard M. Abstract 3797: Chromosome instability as a prognostic factor an immunotherapeutic target in acute myeloid leukemia; Proceedings of the Immunology; Chicago, IL, USA. 14–18 April 2018; p. 3797. American Association for Cancer Research. [Google Scholar]
- 66.Rieder C.L., Cole R.W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V.V.S., Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 68.Kops G.J.P.L., Foltz D.R., Cleveland D.W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K.V., Markowitz S.D., Kinzler K.W., Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 70.Nath S., Ghatak D., Das P., Roychoudhury S. Transcriptional Control of Mitosis: Deregulation and Cancer. Front. Endocrinol. 2015;6 doi: 10.3389/fendo.2015.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X. Correlation of defective mitotic checkpoint with aberrantly reduced expression of MAD2 protein in nasopharyngeal carcinoma cells. Carcinogenesis. 2000;21:2293–2297. doi: 10.1093/carcin/21.12.2293. [DOI] [PubMed] [Google Scholar]
- 72.Hanks S., Coleman K., Reid S., Plaja A., Firth H., FitzPatrick D., Kidd A., Méhes K., Nash R., Robin N., et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 73.Bolanos-Garcia V.M., Blundell T.L. BUB1 and BUBR1: Multifaceted kinases of the cell cycle. Trends Biochem. Sci. 2011;36:141–150. doi: 10.1016/j.tibs.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacquemont S., Bocéno M., Rival J.M., Méchinaud F., David A. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am. J. Med. Genet. 2002;109:17–21. doi: 10.1002/ajmg.10281. [DOI] [PubMed] [Google Scholar]
- 75.Schnerch D., Yalcintepe J., Schmidts A., Becker H., Follo M., Engelhardt M., Wäsch R. Cell cycle control in acute myeloid leukemia. Am. J. Cancer Res. 2012;2:508–528. [PMC free article] [PubMed] [Google Scholar]
- 76.Ghelli Luserna di Rorà A., Martinelli G., Simonetti G. The balance between mitotic death and mitotic slippage in acute leukemia: A new therapeutic window? J. Hematol. Oncol. 2019;12:123. doi: 10.1186/s13045-019-0808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishii K., Usui E., Katayama N., Lorenzo V F., Nakase K., Kobayashi T., Miwa H., Mizutani M., Tanaka I., Nasu K., et al. Characteristics of t(8;21) acute myeloid leukemia (AML) with additional chromosomal abnormality: Concomitant trisomy 4 may constitute a distinctive subtype of t(8;21) AML. Leukemia. 2003;17:731–737. doi: 10.1038/sj.leu.2402871. [DOI] [PubMed] [Google Scholar]
- 78.Boyapati A., Yan M., Peterson L.F., Biggs J.R., Le Beau M.M., Zhang D.-E. A leukemia fusion protein attenuates the spindle checkpoint and promotes aneuploidy. Blood. 2007;109:3963–3971. doi: 10.1182/blood-2006-09-045583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jeganathan K.B., Malureanu L., van Deursen J.M. The Rae1–Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 80.Salsi V., Ferrari S., Gorello P., Fantini S., Chiavolelli F., Mecucci C., Zappavigna V. NUP98 Fusion Oncoproteins Promote Aneuploidy by Attenuating the Mitotic Spindle Checkpoint. Cancer Res. 2014;74:1079–1090. doi: 10.1158/0008-5472.CAN-13-0912. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka K. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cucco F., Musio A. Genome stability: What we have learned from cohesinopathies. Am. J. Med. Genet. Part C Semin. Med. Genet. 2016;172:171–178. doi: 10.1002/ajmg.c.31492. [DOI] [PubMed] [Google Scholar]
- 83.Barber T.D., McManus K., Yuen K.W.Y., Reis M., Parmigiani G., Shen D., Barrett I., Nouhi Y., Spencer F., Markowitz S., et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sajesh B.V., Lichtensztejn Z., McManus K.J. Sister chromatid cohesion defects are associated with chromosome instability in Hodgkin lymphoma cells. BMC Cancer. 2013;13:391. doi: 10.1186/1471-2407-13-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Koninck M., Losada A. Cohesin Mutations in Cancer. Cold Spring Harb. Perspect. Med. 2016;6:a026476. doi: 10.1101/cshperspect.a026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K.J., Kirchner A., Huang L., Chaturvedi A., Wichmann M., et al. Mutations in the cohesin complex in acute myeloid leukemia: Clinical and prognostic implications. Blood. 2014;123:914–920. doi: 10.1182/blood-2013-07-518746. [DOI] [PubMed] [Google Scholar]
- 87.Ostronoff F., Othus M., Lazenby M., Estey E., Appelbaum F.R., Evans A., Godwin J., Gilkes A., Kopecky K.J., Burnett A., et al. Prognostic Significance of NPM1 Mutations in the Absence of FLT3–Internal Tandem Duplication in Older Patients with Acute Myeloid Leukemia: A SWOG and UK National Cancer Research Institute/Medical Research Council Report. J. Clin. Oncol. 2015;33:1157–1164. doi: 10.1200/JCO.2014.58.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim H.-J. Mutations in AML with a normal karyotype: NPM1 and FLT3-ITD, ready to use as a key prognosticator? Korean J. Hematol. 2010;45:79. doi: 10.5045/kjh.2010.45.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cilloni D., Messa F., Rosso V., Arruga F., Defilippi I., Carturan S., Catalano R., Pautasso M., Panuzzo C., Nicoli P., et al. Increase sensitivity to chemotherapeutical agents and cytoplasmatic interaction between NPM leukemic mutant and NF-κB in AML carrying NPM1 mutations. Leukemia. 2008;22:1234–1240. doi: 10.1038/leu.2008.68. [DOI] [PubMed] [Google Scholar]
- 90.Brinkley B. Managing the centrosome numbers game: From chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/S0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 91.Nigg E.A. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 92.Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. Centrosome Abnormalities and Chromosome Instability Occur Together in Pre-invasive Carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 93.Hsu L.-C., Kapali M., DeLoia J.A., Gallion H.H. Centrosome abnormalities in ovarian cancer. Int. J. Cancer. 2005;113:746–751. doi: 10.1002/ijc.20633. [DOI] [PubMed] [Google Scholar]
- 94.Krämer A., Neben K., Ho A.D. Centrosome aberrations in hematological malignancies. Cell Biol. Int. 2005;29:375–383. doi: 10.1016/j.cellbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 95.Pihan G.A. Centrosome Dysfunction Contributes to Chromosome Instability, Chromoanagenesis, and Genome Reprograming in Cancer. Front. Oncol. 2013;3 doi: 10.3389/fonc.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ganem N.J., Godinho S.A., Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Neben K., Giesecke C., Schweizer S., Ho A.D., Krämer A. Centrosome aberrations in acute myeloid leukemia are correlated with cytogenetic risk profile. Blood. 2003;101:289–291. doi: 10.1182/blood-2002-04-1188. [DOI] [PubMed] [Google Scholar]
- 98.Magnaghi-Jaulin L., Eot-Houllier G., Gallaud E., Giet R. Aurora A Protein Kinase: To the Centrosome and Beyond. Biomolecules. 2019;9:28. doi: 10.3390/biom9010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carmena M., Ruchaud S., Earnshaw W.C. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang J., Ikezoe T., Nishioka C., Nobumoto A., Udaka K., Yokoyama A. CD34+/CD38− acute myelogenous leukemia cells aberrantly express Aurora kinase A. Int. J. Cancer. 2013;133:3706–3719. doi: 10.1002/ijc.28277. [DOI] [PubMed] [Google Scholar]
- 101.Ye D., Garcia-Manero G., Kantarjian H.M., Xiao L., Vadhan-Raj S., Fernandez M.H., Nguyen M.H., Medeiros L.J., Bueso-Ramos C.E. Analysis of Aurora kinase A expression in CD34+ blast cells isolated from patients with myelodysplastic syndromes and acute myeloid leukemia. J. Hematop. 2009;2:2–8. doi: 10.1007/s12308-008-0019-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang X.-F., Luo S.-K., Xu J., Li J., Xu D.-R., Wang L.-H., Yan M., Wang X.-R., Wan X.-B., Zheng F.-M., et al. Aurora kinase inhibitory VX-680 increases Bax/Bcl-2 ratio and induces apoptosis in Aurora-A-high acute myeloid leukemia. Blood. 2008;111:2854–2865. doi: 10.1182/blood-2007-07-099325. [DOI] [PubMed] [Google Scholar]
- 103.Lucena-Araujo A.R., de Oliveira F.M., Leite-Cueva S.D., dos Santos G.A., Falcao R.P., Rego E.M. High expression of AURKA and AURKB is associated with unfavorable cytogenetic abnormalities and high white blood cell count in patients with acute myeloid leukemia. Leuk. Res. 2011;35:260–264. doi: 10.1016/j.leukres.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 104.Varga T., Aplan P.D. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair. 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qiu Z., Zhang Z., Roschke A., Varga T., Aplan P.D. Generation of Gross Chromosomal Rearrangements by a Single Engineered DNA Double Strand Break. Sci. Rep. 2017;7:43156. doi: 10.1038/srep43156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ferguson D.O., Alt F.W. DNA double strand break repair and chromosomal translocation: Lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 107.Damelin M., Bestor T.H. The decatenation checkpoint. Br. J. Cancer. 2007;96:201–205. doi: 10.1038/sj.bjc.6603537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khan F.A., Ali S.O. Physiological Roles of DNA Double-Strand Breaks. J. Nucleic Acids. 2017;2017:1–20. doi: 10.1155/2017/6439169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bower J.J., Karaca G.F., Zhou Y., Simpson D.A., Cordeiro-Stone M., Kaufmann W.K. Topoisomerase IIα maintains genomic stability through decatenation G2 checkpoint signaling. Oncogene. 2010;29:4787–4799. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Downes C.S., Clarke D.J., Mullinger A.M., Giménez-Abián J.F., Creighton A.M., Johnson R.T. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 111.Wray J., Williamson E.A., Sheema S., Lee S.-H., Libby E., Willman C.L., Nickoloff J.A., Hromas R. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–1858. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jacoby M.A., De Jesus Pizarro R.E., Shao J., Koboldt D.C., Fulton R.S., Zhou G., Wilson R.K., Walter M.J. The DNA double-strand break response is abnormal in myeloblasts from patients with therapy-related acute myeloid leukemia. Leukemia. 2014;28:1242–1251. doi: 10.1038/leu.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vafa O., Wade M., Kern S., Beeche M., Pandita T.K., Hampton G.M., Wahl G.M. c-Myc Can Induce DNA Damage, Increase Reactive Oxygen Species, and Mitigate p53 Function. Mol. Cell. 2002;9:1031–1044. doi: 10.1016/S1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 114.Kuzyk A., Mai S. c-MYC-Induced Genomic Instability. Cold Spring Harb. Perspect. Med. 2014;4:a014373. doi: 10.1101/cshperspect.a014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karlsson A., Deb-Basu D., Cherry A., Turner S., Ford J., Felsher D.W. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc. Natl. Acad. Sci. USA. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ray S., Atkuri K.R., Deb-Basu D., Adler A.S., Chang H.Y., Herzenberg L.A., Felsher D.W. MYC Can Induce DNA Breaks In vivo and In vitro Independent of Reactive Oxygen Species. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 117.Li Z., Owonikoko T.K., Sun S.-Y., Ramalingam S.S., Doetsch P.W., Xiao Z.-Q., Khuri F.R., Curran W.J., Deng X. c-Myc Suppression of DNA Double-strand Break Repair. Neoplasia. 2012;14:1190–IN35. doi: 10.1593/neo.121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jacobs K.B., Yeager M., Zhou W., Wacholder S., Wang Z., Rodriguez-Santiago B., Hutchinson A., Deng X., Liu C., Horner M.-J., et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Laurie C.C., Laurie C.A., Rice K., Doheny K.F., Zelnick L.R., McHugh C.P., Ling H., Hetrick K.N., Pugh E.W., Amos C., et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grove C.S., Vassiliou G.S. Acute myeloid leukaemia: A paradigm for the clonal evolution of cancer? Dis. Model. Mech. 2014;7:941–951. doi: 10.1242/dmm.015974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hemsing A.L., Hovland R., Tsykunova G., Reikvam H. Trisomy 8 in acute myeloid leukemia. Expert Rev. Hematol. 2019;12:947–958. doi: 10.1080/17474086.2019.1657400. [DOI] [PubMed] [Google Scholar]
- 122.Jaff N., Chelghoum Y., Elhamri M., Tigaud I., Michallet M., Thomas X. Trisomy 8 as sole anomaly or with other clonal aberrations in acute myeloid leukemia: Impact on clinical presentation and outcome. Leuk. Res. 2007;31:67–73. doi: 10.1016/j.leukres.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 123.Vaniawala S.N., Patel M.V., Chavda P.D., Zaveri S.H., Gadhia P.K. The possible significance of trisomy 8 in acute myeloid leukemia. Int. J. Res. Med. Sci. 2017;5:2652. doi: 10.18203/2320-6012.ijrms20172464. [DOI] [Google Scholar]
- 124.De Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 125.De Lange T. How Telomeres Solve the End-Protection Problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Makarov V.L., Hirose Y., Langmore J.P. Long G Tails at Both Ends of Human Chromosomes Suggest a C Strand Degradation Mechanism for Telomere Shortening. Cell. 1997;88:657–666. doi: 10.1016/S0092-8674(00)81908-X. [DOI] [PubMed] [Google Scholar]
- 127.Mattarocci S., D’Ambrosio E., Tafaro L., Somma V., Zannino G., Marigliano V., Ascenzioni F., Cimino-Reale G. Erosion of telomeric 3′-overhangs in white blood cells of aged subjects with high frequency of very short telomeres. Mech. Ageing Dev. 2011;132:27–32. doi: 10.1016/j.mad.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 128.Calado R.T., Regal J.A., Kajigaya S., Young N.S. Erosion of telomeric single-stranded overhang in patients with aplastic anaemia carrying telomerase complex mutations. Eur. J. Clin. Investig. 2009;39:1025–1032. doi: 10.1111/j.1365-2362.2009.02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan S., Han B., Wu Y., Zhou D., Zhao Y. Telomerase gene mutation screening and telomere overhang detection in Chinese patients with acute myeloid leukemia. Leuk. Lymphoma. 2013;54:1437–1441. doi: 10.3109/10428194.2012.729834. [DOI] [PubMed] [Google Scholar]
- 130.Bernadotte A., Mikhelson V.M., Spivak I.M. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging. 2016;8:3–11. doi: 10.18632/aging.100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 132.Scheel C., Schaefer K.-L., Jauch A., Keller M., Wai D., Brinkschmidt C., van Valen F., Boecker W., Dockhorn-Dworniczak B., Poremba C. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene. 2001;20:3835–3844. doi: 10.1038/sj.onc.1204493. [DOI] [PubMed] [Google Scholar]
- 133.Jafri M.A., Ansari S.A., Alqahtani M.H., Shay J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8:69. doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aalbers A.M., Calado R.T., Young N.S., Zwaan C.M., Wu C., Kajigaya S., Coenen E.A., Baruchel A., Geleijns K., de Haas V., et al. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia. 2013;27:1786–1789. doi: 10.1038/leu.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mosrati M.A., Willander K., Falk I.J., Hermanson M., Höglund M., Stockelberg D., Wei Y., Lotfi K., Söderkvist P. Association between TERT promoter polymorphisms and acute myeloid leukemia risk and prognosis. Oncotarget. 2015;6:25109–25120. doi: 10.18632/oncotarget.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Swiggers S.J.J., Kuijpers M.A., de Cort M.J.M., Beverloo H.B., Zijlmans J.M.J.M. Critically short telomeres in acute myeloid leukemia with loss or gain of parts of chromosomes. Genes Chromosom. Cancer. 2006;45:247–256. doi: 10.1002/gcc.20286. [DOI] [PubMed] [Google Scholar]
- 137.Hartmann U., Brümmendorf T.H., Balabanov S., Thiede C., Illme T., Schaich M. Telomere length and hTERT expression in patients with acute myeloid leukemia correlates with chromosomal abnormalities. Haematologica. 2005;90:307–316. [PubMed] [Google Scholar]
- 138.Capraro V., Zane L., Poncet D., Perol D., Galia P., Preudhomme C., Bonnefoy-Berard N., Gilson E., Thomas X., El-Hamri M. Telomere deregulations possess cytogenetic, phenotype, and prognostic specificities in acute leukemias. Exp. Hematol. 2011;39:195–202.e2. doi: 10.1016/j.exphem.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 139.Alter B.P., Giri N., Savage S.A., Rosenberg P.S. Cancer in dyskeratosis congenita. Blood. 2009;113:6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gadji M., Adebayo Awe J., Rodrigues P., Kumar R., Houston D.S., Klewes L., Dieye T.N., Rego E.M., Passetto R.F., de Oliveira F.M., et al. Profiling Three-Dimensional Nuclear Telomeric Architecture of Myelodysplastic Syndromes and Acute Myeloid Leukemia Defines Patient Subgroups. Clin. Cancer Res. 2012;18:3293–3304. doi: 10.1158/1078-0432.CCR-12-0087. [DOI] [PubMed] [Google Scholar]
- 141.Sieglová Z., Žilovcová S., Čermák J., Říhová H., Březinová D., Dvořáková R., Marková M., Maaloufová J., Sajdová J., Březinová J., et al. Dynamics of telomere erosion and its association with genome instability in myelodysplastic syndromes (MDS) and acute myelogenous leukemia arising from MDS: A marker of disease prognosis? Leuk. Res. 2004;28:1013–1021. doi: 10.1016/j.leukres.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 142.Warny M., Helby J., Sengeløv H., Nordestgaard B.G., Birgens H., Bojesen S.E. Bone marrow mononuclear cell telomere length in acute myeloid leukaemia and high-risk myelodysplastic syndrome. Eur. J. Haematol. 2019;102:218–226. doi: 10.1111/ejh.13196. [DOI] [PubMed] [Google Scholar]
- 143.XU D., Gruber A., Peterson C., Pisa P. Telomerase activity and the expression of telomerase components in acute myelogenous leukaemia. Br. J. Haematol. 1998;102:1367–1375. doi: 10.1046/j.1365-2141.1998.00969.x. [DOI] [PubMed] [Google Scholar]
- 144.Van Steensel B., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 145.Vajen B., Thomay K., Schlegelberger B. Induction of Chromosomal Instability via Telomere Dysfunction and Epigenetic Alterations in Myeloid Neoplasia. Cancers. 2013;5:857–874. doi: 10.3390/cancers5030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mai S., Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J. Cell. Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 147.Calado R.T., Cooper J.N., Padilla-Nash H.M., Sloand E.M., Wu C.O., Scheinberg P., Ried T., Young N.S. Short telomeres result in chromosomal instability in hematopoietic cells and precede malignant evolution in human aplastic anemia. Leukemia. 2012;26:700–707. doi: 10.1038/leu.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Louis S.F., Vermolen B.J., Garini Y., Young I.T., Guffei A., Lichtensztejn Z., Kuttler F., Chuang T.C.Y., Moshir S., Mougey V., et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc. Natl. Acad. Sci. USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Andersen M., Pedersen-Bjergaard J. Increased frequency of dicentric chromosomes in therapy-related MDS and AML compared to de novo disease is significantly related to previous treatment with alkylating agents and suggests a specific susceptibility to chromosome breakage at the centromere. Leukemia. 2000;14:105–111. doi: 10.1038/sj.leu.2401594. [DOI] [PubMed] [Google Scholar]
- 150.MacKinnon R.N., Campbell L.J. The Role of Dicentric Chromosome Formation and Secondary Centromere Deletion in the Evolution of Myeloid Malignancy. Genet. Res. Int. 2011;2011:1–11. doi: 10.4061/2011/643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sarova I., Brezinova J., Zemanova Z., Ransdorfova S., Izakova S., Svobodova K., Pavlistova L., Berkova A., Cermak J., Jonasova A., et al. Molecular cytogenetic analysis of dicentric chromosomes in acute myeloid leukemia. Leuk. Res. 2016;43:51–57. doi: 10.1016/j.leukres.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 152.Gascoigne K.E., Cheeseman I.M. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosom. Res. 2013;21:407–418. doi: 10.1007/s10577-013-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rausch T., Jones D.T.W., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A., et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bochtler T., Granzow M., Stölzel F., Kunz C., Mohr B., Kartal-Kaess M., Hinderhofer K., Heilig C.E., Kramer M., Thiede C., et al. Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia. Blood. 2017;129:1333–1342. doi: 10.1182/blood-2016-09-738161. [DOI] [PubMed] [Google Scholar]
- 155.Fontana M.C., Marconi G., Feenstra J.D.M., Fonzi E., Papayannidis C., Ghelli Luserna di Rorá A., Padella A., Solli V., Franchini E., Ottaviani E., et al. Chromothripsis in acute myeloid leukemia: Biological features and impact on survival. Leukemia. 2018;32:1609–1620. doi: 10.1038/s41375-018-0035-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Koduru P.R., Wilson K., Wen J., Garcia R., Patel S., Monaghan S.A. Cytogenetic and Cytogenomic Microarray Characterization of Chromothripsis in Chromosome 8 Affecting MOZ/NCOA2 (TIF2), FGFR1, RUNX1T1, and RUNX1 in a Pediatric Acute Myeloid Leukemia. J. Pediatr. Hematol. Oncol. 2017;39:e227–e232. doi: 10.1097/MPH.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 157.Righolt C., Mai S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosom. Cancer. 2012;51:975–981. doi: 10.1002/gcc.21981. [DOI] [PubMed] [Google Scholar]
- 158.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rücker F.G., Dolnik A., Blätte T.J., Teleanu V., Ernst A., Thol F., Heuser M., Ganser A., Döhner H., Döhner K., et al. Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype. Haematologica. 2018;103:e17–e20. doi: 10.3324/haematol.2017.180497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gao J., Chen Y.-H., Mina A., Altman J.K., Kim K.-Y., Zhang Y., Lu X., Jennings L., Sukhanova M. Unique morphologic and genetic characteristics of acute myeloid leukemia with chromothripsis: A clinicopathologic study from a single institution. Hum. Pathol. 2020;98:22–31. doi: 10.1016/j.humpath.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 161.L′Abbate A., Tolomeo D., Cifola I., Severgnini M., Turchiano A., Augello B., Squeo G., D′Addabbo P., Traversa D., Daniele G., et al. MYC-containing amplicons in acute myeloid leukemia: Genomic structures, evolution, and transcriptional consequences. Leukemia. 2018;32:2152–2166. doi: 10.1038/s41375-018-0033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Descartes M., Korf B.R., Mikhail F.M. Swaiman’s Pediatric Neurology. Elsevier; Amsterdam, The Netherlands: 2017. Chromosomes and Chromosomal Abnormalities; pp. 268–276. [Google Scholar]
- 163.Fuse K., Tanaka T., Shibasaki Y., Furukawa T., Narita M., Sone H., Masuko M. Marker chromosome is a strong poor prognosis factor after allogeneic HSCT for adverse-risk AML patients. Eur. J. Haematol. 2020:ejh.13495. doi: 10.1111/ejh.13495. [DOI] [PubMed] [Google Scholar]
- 164.Liu W.-J., Tan X.-H., Luo X.-P., Guo B.-P., Wei Z.-J., Ke Q., He S., Cen H. Prognostic significance of Tet methylcytosine dioxygenase 2 (TET2) gene mutations in adult patients with acute myeloid leukemia: A meta-analysis. Leuk. Lymphoma. 2014;55:2691–2698. doi: 10.3109/10428194.2014.893308. [DOI] [PubMed] [Google Scholar]
- 165.Xu F., Li X., Wu L., Zhang Q., Yang R., Yang Y., Zhang Z., He Q., Chang C. Overexpression of the EZH2, RING1 and BMI1 genes is common in myelodysplastic syndromes: Relation to adverse epigenetic alteration and poor prognostic scoring. Ann. Hematol. 2011;90:643–653. doi: 10.1007/s00277-010-1128-5. [DOI] [PubMed] [Google Scholar]
- 166.Sun Y., Chen B.-R., Deshpande A. Epigenetic Regulators in the Development, Maintenance, and Therapeutic Targeting of Acute Myeloid Leukemia. Front. Oncol. 2018;8 doi: 10.3389/fonc.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kishtagari A., Levine R.L., Viny A.D. Driver mutations in acute myeloid leukemia. Curr. Opin. Hematol. 2020;27:49–57. doi: 10.1097/MOH.0000000000000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Desai P., Mencia-Trinchant N., Savenkov O., Simon M.S., Cheang G., Lee S., Samuel M., Ritchie E.K., Guzman M.L., Ballman K.V., et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 2018;24:1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Abelson S., Collord G., Ng S.W.K., Weissbrod O., Mendelson Cohen N., Niemeyer E., Barda N., Zuzarte P.C., Heisler L., Sundaravadanam Y., et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Döhner K., Brown J., Hehmann U., Hetzel C., Stewart J., Lowther G., Scholl C., Fröhling S., Cuneo A., Tsui L.C., et al. Molecular Cytogenetic Characterization of a Critical Region in Bands 7q35-q36 Commonly Deleted in Malignant Myeloid Disorders. Blood. 1998;92:4031–4035. doi: 10.1182/blood.V92.11.4031. [DOI] [PubMed] [Google Scholar]
- 171.Jerez A., Sugimoto Y., Makishima H., Verma A., Jankowska A.M., Przychodzen B., Visconte V., Tiu R.V., O’Keefe C.L., Mohamedali A.M., et al. Loss of heterozygosity in 7q myeloid disorders: Clinical associations and genomic pathogenesis. Blood. 2012;119:6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mrózek K., Eisfeld A.-K., Kohlschmidt J., Carroll A.J., Walker C.J., Nicolet D., Blachly J.S., Bill M., Papaioannou D., Wang E.S., et al. Complex karyotype in de novo acute myeloid leukemia: Typical and atypical subtypes differ molecularly and clinically. Leukemia. 2019;33:1620–1634. doi: 10.1038/s41375-019-0390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang J., He N., Wang R., Tian T., Han F., Zhong C., Zhang C., Hua M., Ji C., Ma D. Analysis of TET2 and EZH2 gene functions in chromosome instability in acute myeloid leukemia. Sci. Rep. 2020;10:2706. doi: 10.1038/s41598-020-59365-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schvartzman J.-M., Duijf P.H.G., Sotillo R., Coker C., Benezra R. Mad2 Is a Critical Mediator of the Chromosome Instability Observed upon Rb and p53 Pathway Inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sotillo R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., Benezra R. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mondal G., Sengupta S., Panda C.K., Gollin S.M., Saunders W.S., Roychoudhury S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28:81–92. doi: 10.1093/carcin/bgl100. [DOI] [PubMed] [Google Scholar]
- 177.Simonetti G., Padella A., do Valle I.F., Fontana M.C., Fonzi E., Bruno S., Baldazzi C., Guadagnuolo V., Manfrini M., Ferrari A., et al. Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer. 2019;125:712–725. doi: 10.1002/cncr.31837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen Y., Kantarjian H., Wang H., Cortes J., Ravandi F. Acute promyelocytic leukemia: A population-based study on incidence and survival in the United States, 1975–2008. Cancer. 2012;118:5811–5818. doi: 10.1002/cncr.27623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Labrador J., Luño E., Vellenga E., Brunet S., González-Campos J., Chillón M.C., Holowiecka A., Esteve J., Bergua J., González-Sanmiguel J.D., et al. Clinical significance of complex karyotype at diagnosis in pediatric and adult patients with de novo acute promyelocytic leukemia treated with ATRA and chemotherapy. Leuk. Lymphoma. 2019;60:1146–1155. doi: 10.1080/10428194.2018.1522438. [DOI] [PubMed] [Google Scholar]
- 180.Lushnikova T., Bouska A., Odvody J., Dupont W.D., Eischen C.M. Aging mice have increased chromosome instability that is exacerbated by elevated Mdm2 expression. Oncogene. 2011;30:4622–4631. doi: 10.1038/onc.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hellmich C., Moore J.A., Bowles K.M., Rushworth S.A. Bone Marrow Senescence and the Microenvironment of Hematological Malignancies. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vaziri H., Dragowska W., Allsopp R.C., Thomas T.E., Harley C.B., Lansdorp P.M. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Silva P., Neumann M., Schroeder M.P., Vosberg S., Schlee C., Isaakidis K., Ortiz-Tanchez J., Fransecky L.R., Hartung T., Türkmen S., et al. Acute myeloid leukemia in the elderly is characterized by a distinct genetic and epigenetic landscape. Leukemia. 2017;31:1640–1644. doi: 10.1038/leu.2017.109. [DOI] [PubMed] [Google Scholar]
- 184.Acuna-Hidalgo R., Sengul H., Steehouwer M., van de Vorst M., Vermeulen S.H., Kiemeney L.A.L.M., Veltman J.A., Gilissen C., Hoischen A. Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life. Am. J. Hum. Genet. 2017;101:50–64. doi: 10.1016/j.ajhg.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Granfeldt Østgård L.S., Medeiros B.C., Sengeløv H., Nørgaard M., Andersen M.K., Dufva I.H., Friis L.S., Kjeldsen E., Marcher C.W., Preiss B., et al. Epidemiology and Clinical Significance of Secondary and Therapy-Related Acute Myeloid Leukemia: A National Population-Based Cohort Study. J. Clin. Oncol. 2015;33:3641–3649. doi: 10.1200/JCO.2014.60.0890. [DOI] [PubMed] [Google Scholar]
- 186.Kern W., Haferlach T., Schnittger S., Ludwig W., Hiddemann W., Schoch C. Karyotype instability between diagnosis and relapse in 117 patients with acute myeloid leukemia: Implications for resistance against therapy. Leukemia. 2002;16:2084–2091. doi: 10.1038/sj.leu.2402654. [DOI] [PubMed] [Google Scholar]
- 187.Feurstein S., Thomay K., Hofmann W., Buesche G., Kreipe H., Thol F., Heuser M., Ganser A., Schlegelberger B., Göhring G. Routes of Clonal Evolution into Complex Karyotypes in Myelodysplastic Syndrome Patients with 5q Deletion. Int. J. Mol. Sci. 2018;19:3269. doi: 10.3390/ijms19103269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moison C., Lavallée V.-P., Thiollier C., Lehnertz B., Boivin I., Mayotte N., Gareau Y., Fréchette M., Blouin-Chagnon V., Corneau S., et al. Complex karyotype AML displays G2/M signature and hypersensitivity to PLK1 inhibition. Blood Adv. 2019;3:552–563. doi: 10.1182/bloodadvances.2018028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Leung G.M.K., Zhang C., Ng N.K.L., Yang N., Lam S.S.Y., Au C.H., Chan T.L., Ma E.S.K., Tsui S.P., Ip H.W., et al. Distinct mutation spectrum, clinical outcome and therapeutic responses of typical complex/monosomy karyotype acute myeloid leukemia carrying TP53 mutations. Am. J. Hematol. 2019;94:650–657. doi: 10.1002/ajh.25469. [DOI] [PubMed] [Google Scholar]
- 190.Parkin B., Ouillette P., Li Y., Keller J., Lam C., Roulston D., Li C., Shedden K., Malek S.N. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. 2013;121:369–377. doi: 10.1182/blood-2012-04-427039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bochtler T., Fröhling S., Krämer A. Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia. Leukemia. 2015;29:1243–1252. doi: 10.1038/leu.2015.32. [DOI] [PubMed] [Google Scholar]
- 192.Hou H.-A., Chou W.-C., Kuo Y.-Y., Liu C.-Y., Lin L.-I., Tseng M.-H., Chiang Y.-C., Liu M.-C., Liu C.-W., Tang J.-L., et al. TP53 mutations in de novo acute myeloid leukemia patients: Longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J. 2015;5:e331. doi: 10.1038/bcj.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wong T.N., Ramsingh G., Young A.L., Miller C.A., Touma W., Welch J.S., Lamprecht T.L., Shen D., Hundal J., Fulton R.S., et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cluzeau T., Sebert M., Rahmé R., Cuzzubbo S., Walter-petrich A., Lehmann che J., Peterlin P., Beve B., Attalah H., Chermat F., et al. APR-246 Combined with Azacitidine (AZA) in TP53 Mutated Myelodysplastic Syndrome (MDS) and Acute Myeloid Leukemia (AML). a Phase 2 Study By the Groupe Francophone Des Myélodysplasies (GFM) Blood. 2019;134:677. doi: 10.1182/blood-2019-125579. [DOI] [Google Scholar]
- 196.Gnyszka A., Jastrzebski Z., Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33:2989–2996. [PubMed] [Google Scholar]
- 197.Sallman D.A., DeZern A.E., Garcia-Manero G., Steensma D.P., Roboz G.J., Sekeres M.A., Cluzeau T., Sweet K.L., McLemore A.F., McGraw K., et al. Phase 2 Results of APR-246 and Azacitidine (AZA) in Patients with TP53 mutant Myelodysplastic Syndromes (MDS) and Oligoblastic Acute Myeloid Leukemia (AML) Blood. 2019;134:676. doi: 10.1182/blood-2019-131055. [DOI] [Google Scholar]
- 198.Sallman D.A., DeZern A.E., Garcia-Manero G., Steensma D.P., Roboz G.J., Sekeres M.A., Cluzeau T., Sweet K.L., McLemore A., McGraw K.L., et al. Eprenetapopt (APR-246) and Azacitidine in TP53 -Mutant Myelodysplastic Syndromes. J. Clin. Oncol. 2021;39:1584–1594. doi: 10.1200/JCO.20.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Göhring G., Giagounidis A., Büsche G., Kreipe H.H., Zimmermann M., Hellström-Lindberg E., Aul C., Schlegelberger B. Patients with del(5q) MDS who fail to achieve sustained erythroid or cytogenetic remission after treatment with lenalidomide have an increased risk for clonal evolution and AML progression. Ann. Hematol. 2010;89:365–374. doi: 10.1007/s00277-009-0846-z. [DOI] [PubMed] [Google Scholar]
- 200.Middeke J.M., Beelen D., Stadler M., Göhring G., Schlegelberger B., Baurmann H., Bug G., Bellos F., Mohr B., Buchholz S., et al. Outcome of high-risk acute myeloid leukemia after allogeneic hematopoietic cell transplantation: Negative impact of abnl(17p) and −5/5q−. Blood. 2012;120:2521–2528. doi: 10.1182/blood-2012-03-417972. [DOI] [PubMed] [Google Scholar]
- 201.Bulaeva E., Pellacani D., Nakamichi N., Hammond C.A., Beer P.A., Lorzadeh A., Moksa M., Carles A., Bilenky M., Lefort S., et al. MYC-induced human acute myeloid leukemia requires a continuing IL-3/GM-CSF costimulus. Blood. 2020;136:2764–2773. doi: 10.1182/blood.2020006374. [DOI] [PubMed] [Google Scholar]
- 202.Pan X.-N., Chen J.-J., Wang L.-X., Xiao R.-Z., Liu L.-L., Fang Z.-G., Liu Q., Long Z.-J., Lin D.-J. Inhibition of c-Myc Overcomes Cytotoxic Drug Resistance in Acute Myeloid Leukemia Cells by Promoting Differentiation. PLoS ONE. 2014;9:e105381. doi: 10.1371/journal.pone.0105381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mertz J.A., Conery A.R., Bryant B.M., Sandy P., Balasubramanian S., Mele D.A., Bergeron L., Sims R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dawson M., Stein E.M., Huntly B.J.P., Karadimitris A., Kamdar M., Fernandez de Larrea C., Dickinson M.J., Yeh P.S.-H., Daver N., Chaidos A., et al. A Phase I Study of GSK525762, a Selective Bromodomain (BRD) and Extra Terminal Protein (BET) Inhibitor: Results from Part 1 of Phase I/II Open Label Single Agent Study in Patients with Acute Myeloid Leukemia (AML) Blood. 2017;130:1377. doi: 10.1182/blood.V130.Suppl_1.1377.1377. [DOI] [Google Scholar]
- 205.Kang C., Kim C.-Y., Kim H.S., Park S.-P., Chung H.-M. The Bromodomain Inhibitor JQ1 Enhances the Responses to All- trans Retinoic Acid in HL-60 and MV4-11 Leukemia Cells. Int. J. Stem Cells. 2018;11:131–140. doi: 10.15283/ijsc18021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Call S.G., Duren R.P., Panigrahi A.K., Nguyen L., Freire P.R., Grimm S.L., Coarfa C., Conneely O.M. Targeting Oncogenic Super Enhancers in MYC-Dependent AML Using a Small Molecule Activator of NR4A Nuclear Receptors. Sci. Rep. 2020;10:2851. doi: 10.1038/s41598-020-59469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Brunner A.M., Blonquist T.M., DeAngelo D.J., McMasters M., Winer E.S., Hobbs G.S., Amrein P.C., Hock H., Steensma D.P., Garcia J.S., et al. Phase II Clinical Trial of Alisertib, an Aurora a Kinase Inhibitor, in Combination with Induction Chemotherapy in High-Risk, Untreated Patients with Acute Myeloid Leukemia. Blood. 2018;132:766. doi: 10.1182/blood-2018-99-115145. [DOI] [Google Scholar]
- 208.Sarova I., Brezinova J., Zemanova Z., Ransdorfova S., Svobodova K., Izakova S., Pavlistova L., Lizcova L., Berkova A., Skipalova K., et al. High frequency of dicentric chromosomes detected by multi-centromeric FISH in patients with acute myeloid leukemia and complex karyotype. Leuk. Res. 2018;68:85–89. doi: 10.1016/j.leukres.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 209.Goldenson B., Crispino J.D. The aurora kinases in cell cycle and leukemia. Oncogene. 2015;34:537–545. doi: 10.1038/onc.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Goldberg S.L., Fenaux P., Craig M.D., Gyan E., Lister J., Kassis J., Pigneux A., Schiller G.J., Jung J., Jane Leonard E., et al. An exploratory phase 2 study of investigational Aurora A kinase inhibitor alisertib (MLN8237) in acute myelogenous leukemia and myelodysplastic syndromes. Leuk. Res. Rep. 2014;3:58–61. doi: 10.1016/j.lrr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fathi A.T., Wander S.A., Blonquist T.M., Brunner A.M., Amrein P.C., Supko J., Hermance N.M., Manning A.L., Sadrzadeh H., Ballen K.K., et al. Phase I study of the aurora A kinase inhibitor alisertib with induction chemotherapy in patients with acute myeloid leukemia. Haematologica. 2017;102:719–727. doi: 10.3324/haematol.2016.158394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.DiNardo C.D., Jonas B.A., Pullarkat V., Thirman M.J., Garcia J.S., Wei A.H., Konopleva M., Döhner H., Letai A., Fenaux P., et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N. Engl. J. Med. 2020;383:617–629. doi: 10.1056/NEJMoa2012971. [DOI] [PubMed] [Google Scholar]
- 213.Payton M., Cheung H.-K., Ninniri M.S.S., Marinaccio C., Wayne W.C., Hanestad K., Crispino J.D., Juan G., Coxon A. Dual Targeting of Aurora Kinases with AMG 900 Exhibits Potent Preclinical Activity Against Acute Myeloid Leukemia with Distinct Post-Mitotic Outcomes. Mol. Cancer Ther. 2018;17:2575–2585. doi: 10.1158/1535-7163.MCT-18-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Qi J., Gao X., Zhong X., Zhang N., Wang R., Zhang H., Pan T., Liu X., Yao Y., Wu Q., et al. Selective inhibition of Aurora A and B kinases effectively induces cell cycle arrest in t(8;21) acute myeloid leukemia. Biomed. Pharmacother. 2019;117:109113. doi: 10.1016/j.biopha.2019.109113. [DOI] [PubMed] [Google Scholar]
- 215.Dennis M., Davies M., Oliver S., D’Souza R., Pike L., Stockman P. Phase I study of the Aurora B kinase inhibitor barasertib (AZD1152) to assess the pharmacokinetics, metabolism and excretion in patients with acute myeloid leukemia. Cancer Chemother. Pharmacol. 2012;70:461–469. doi: 10.1007/s00280-012-1939-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Löwenberg B., Muus P., Ossenkoppele G., Rousselot P., Cahn J.-Y., Ifrah N., Martinelli G., Amadori S., Berman E., Sonneveld P., et al. Phase 1/2 study to assess the safety, efficacy, and pharmacokinetics of barasertib (AZD1152) in patients with advanced acute myeloid leukemia. Blood. 2011;118:6030–6036. doi: 10.1182/blood-2011-07-366930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tsuboi K., Yokozawa T., Sakura T., Watanabe T., Fujisawa S., Yamauchi T., Uike N., Ando K., Kihara R., Tobinai K., et al. A Phase I study to assess the safety, pharmacokinetics and efficacy of barasertib (AZD1152), an Aurora B kinase inhibitor, in Japanese patients with advanced acute myeloid leukemia. Leuk. Res. 2011;35:1384–1389. doi: 10.1016/j.leukres.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 218.Kantarjian H.M., Sekeres M.A., Ribrag V., Rousselot P., Garcia-Manero G., Jabbour E.J., Owen K., Stockman P.K., Oliver S.D. Phase I Study Assessing the Safety and Tolerability of Barasertib (AZD1152) With Low-Dose Cytosine Arabinoside in Elderly Patients With AML. Clin. Lymphoma Myeloma Leuk. 2013;13:559–567. doi: 10.1016/j.clml.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hartsink-Segers S.A., Zwaan C.M., Exalto C., Luijendijk M.W.J., Calvert V.S., Petricoin E.F., Evans W.E., Reinhardt D., de Haas V., Hedtjärn M., et al. Aurora kinases in childhood acute leukemia: The promise of aurora B as therapeutic target. Leukemia. 2013;27:560–568. doi: 10.1038/leu.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ikezoe T., Yang J., Nishioka C., Yokoyama A. p53 is critical for the Aurora B kinase inhibitor-mediated apoptosis in acute myelogenous leukemia cells. Int. J. Hematol. 2009;91:69–77. doi: 10.1007/s12185-009-0462-7. [DOI] [PubMed] [Google Scholar]
- 221.Quivoron C., Couronné L., Della Valle V., Lopez C.K., Plo I., Wagner-Ballon O., Do Cruzeiro M., Delhommeau F., Arnulf B., Stern M.-H., et al. TET2 Inactivation Results in Pleiotropic Hematopoietic Abnormalities in Mouse and Is a Recurrent Event during Human Lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 222.Chou W.-C., Chou S.-C., Liu C.-Y., Chen C.-Y., Hou H.-A., Kuo Y.-Y., Lee M.-C., Ko B.-S., Tang J.-L., Yao M., et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. 2011;118:3803–3810. doi: 10.1182/blood-2011-02-339747. [DOI] [PubMed] [Google Scholar]
- 223.Cimmino L., Dolgalev I., Wang Y., Yoshimi A., Martin G.H., Wang J., Ng V., Xia B., Witkowski M.T., Mitchell-Flack M., et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell. 2017;170:1079–1095.e20. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Das A.B., Kakadia P.M., Wojcik D., Pemberton L., Browett P.J., Bohlander S.K., Vissers M.C.M. Clinical remission following ascorbate treatment in a case of acute myeloid leukemia with mutations in TET2 and WT1. Blood Cancer J. 2019;9:82. doi: 10.1038/s41408-019-0242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Göllner S., Oellerich T., Agrawal-Singh S., Schenk T., Klein H.-U., Rohde C., Pabst C., Sauer T., Lerdrup M., Tavor S., et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 2017;23:69–78. doi: 10.1038/nm.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kayser S., Döhner K., Krauter J., Köhne C.-H., Horst H.A., Held G., von Lilienfeld-Toal M., Wilhelm S., Kündgen A., Götze K., et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood. 2011;117:2137–2145. doi: 10.1182/blood-2010-08-301713. [DOI] [PubMed] [Google Scholar]
- 227.Smith S.M., Le Beau M.M., Huo D., Karrison T., Sobecks R.M., Anastasi J., Vardiman J.W., Rowley J.D., Larson R.A. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: The University of Chicago series. Blood. 2003;102:43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 228.Byrd J., Mrózek K., Dodge R., Carroll A., Edwards C., Arthur D., Pettenati M., Patil S., Rao K., Watson M., et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 229.Lindsley R.C., Mar B.G., Mazzola E., Grauman P.V., Shareef S., Allen S.L., Pigneux A., Wetzler M., Stuart R.K., Erba H.P., et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Schoch C., Kern W., Kohlmann A., Hiddemann W., Schnittger S., Haferlach T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosom. Cancer. 2005;43:227–238. doi: 10.1002/gcc.20193. [DOI] [PubMed] [Google Scholar]
- 231.Watts J.M., Dumitriu B., Hilden P., Chen C., Rapaport F., Kishtagari A., Ahn J., Devlin S.M., Rampal R.K., Levine R.L., et al. Telomere Length Is Associated with Specific Mutations and Mutation Classes in Patients with Acute Myeloid Leukemia. Blood. 2014;124:2280. doi: 10.1182/blood.V124.21.2280.2280. [DOI] [Google Scholar]
- 232.Ohanian M., Rozovski U., Kanagal-Shamanna R., Abruzzo L.V., Loghavi S., Kadia T., Futreal A., Bhalla K., Zuo Z., Huh Y.O., et al. MYC protein expression is an important prognostic factor in acute myeloid leukemia. Leuk. Lymphoma. 2019;60:37–48. doi: 10.1080/10428194.2018.1464158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Koh Y., Kim I., Bae J.-Y., Song E.Y., Kim H.-K., Yoon S.-S., Lee D.S., Park S.S., Park M.H., Park S., et al. Prognosis of Secondary Acute Myeloid Leukemia is Affected by the Type of the Preceding Hematologic Disorders and the Presence of Trisomy 8. Jpn. J. Clin. Oncol. 2010;40:1037–1045. doi: 10.1093/jjco/hyq097. [DOI] [PubMed] [Google Scholar]
- 234.Weissmann S., Alpermann T., Grossmann V., Kowarsch A., Nadarajah N., Eder C., Dicker F., Fasan A., Haferlach C., Haferlach T., et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26:934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 235.Delhommeau F., Dupont S., Valle V.D., James C., Trannoy S., Massé A., Kosmider O., Le Couedic J.-P., Robert F., Alberdi A., et al. Mutation in TET2 in Myeloid Cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 236.Wang X., Dai H., Wang Q., Wang Q., Xu Y., Wang Y., Sun A., Ruan J., Chen S., Wu D. EZH2 Mutations Are Related to Low Blast Percentage in Bone Marrow and -7/del(7q) in De Novo Acute Myeloid Leukemia. PLoS ONE. 2013;8:e61341. doi: 10.1371/journal.pone.0061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stasik S., Middeke J.M., Kramer M., Röllig C., Krämer A., Scholl S., Hochhaus A., Crysandt M., Brümmendorf T.H., Naumann R., et al. EZH2 mutations and impact on clinical outcome: An analysis in 1604 patients with newly diagnosed acute myeloid leukemia. Haematologica. 2020;105:e228–e231. doi: 10.3324/haematol.2019.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kelly K.R., Nawrocki S.T., Espitia C.M., Zhang M., Yang J.J., Padmanabhan S., Ecsedy J., Giles F.J., Carew J.S. Targeting Aurora A kinase activity with the investigational agent alisertib increases the efficacy of cytarabine through a FOXO-dependent mechanism. Int. J. Cancer. 2012;131:2693–2703. doi: 10.1002/ijc.27579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wen Q., Goldenson B., Silver S.J., Schenone M., Dancik V., Huang Z., Wang L.-Z., Lewis T.A., An W.F., Li X., et al. Identification of Regulators of Polyploidization Presents Therapeutic Targets for Treatment of AMKL. Cell. 2012;150:575–589. doi: 10.1016/j.cell.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Krause D.S., Crispino J.D. Molecular Pathways: Induction of Polyploidy as a Novel Differentiation Therapy for Leukemia. Clin. Cancer Res. 2013;19:6084–6088. doi: 10.1158/1078-0432.CCR-12-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bruedigam C., Bagger F.O., Heidel F.H., Paine Kuhn C., Guignes S., Song A., Austin R., Vu T., Lee E., Riyat S., et al. Telomerase Inhibition Effectively Targets Mouse and Human AML Stem Cells and Delays Relapse following Chemotherapy. Cell Stem Cell. 2014;15:775–790. doi: 10.1016/j.stem.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hidaka D., Onozawa M., Miyashita N., Yokoyama S., Nakagawa M., Hashimoto D., Teshima T. Short-term treatment with imetelstat sensitizes hematopoietic malignant cells to a genotoxic agent via suppression of the telomerase-mediated DNA repair process. Leuk. Lymphoma. 2020;61:2722–2732. doi: 10.1080/10428194.2020.1779256. [DOI] [PubMed] [Google Scholar]
- 244.Rusbuldt J.J., Luistro L., Chin D., Smith M., Wong A., Romero M., Rizo A., Bussolari J., Huang F., Sasser A. (Kate) Abstract 1101: Telomerase inhibitor imetelstat in combination with the BCL-2 inhibitor venetoclax enhances apoptosis in vitro and increases survival in vivo in acute myeloid leukemia; Proceedings of the Experimental and Molecular Therapeutics; Washington, DC, USA. 1–5 April 2017; Philadelphia, PA, USA: American Association for Cancer Research; 2017. p. 1101. [Google Scholar]
- 245.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., et al. BET Bromodomain Inhibition as a Therapeutic Strategy to Target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Agathocleous M., Meacham C.E., Burgess R.J., Piskounova E., Zhao Z., Crane G.M., Cowin B.L., Bruner E., Murphy M.M., Chen W., et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549:476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Vissers M.C.M., Das A.B. Potential Mechanisms of Action for Vitamin C in Cancer: Reviewing the Evidence. Front. Physiol. 2018;9 doi: 10.3389/fphys.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mingay M., Chaturvedi A., Bilenky M., Cao Q., Jackson L., Hui T., Moksa M., Heravi-Moussavi A., Humphries R.K., Heuser M., et al. Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia. 2018;32:11–20. doi: 10.1038/leu.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zhao H., Zhu H., Huang J., Zhu Y., Hong M., Zhu H., Zhang J., Li S., Yang L., Lian Y., et al. The synergy of Vitamin C with decitabine activates TET2 in leukemic cells and significantly improves overall survival in elderly patients with acute myeloid leukemia. Leuk. Res. 2018;66:1–7. doi: 10.1016/j.leukres.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 250.Renner A.G., Dos Santos C., Recher C., Bailly C., Créancier L., Kruczynski A., Payrastre B., Manenti S. Polo-like kinase 1 is overexpressed in acute myeloid leukemia and its inhibition preferentially targets the proliferation of leukemic cells. Blood. 2009;114:659–662. doi: 10.1182/blood-2008-12-195867. [DOI] [PubMed] [Google Scholar]
- 251.Cunningham C.E., MacAuley M.J., Vizeacoumar F.S., Abuhussein O., Freywald A., Vizeacoumar F.J. The CINs of Polo-Like Kinase 1 in Cancer. Cancers. 2020;12:2953. doi: 10.3390/cancers12102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ikezoe T., Yang J., Nishioka C., Takezaki Y., Tasaka T., Togitani K., Koeffler H.P., Yokoyama A. A novel treatment strategy targeting polo-like kinase 1 in hematological malignancies. Leukemia. 2009;23:1564–1576. doi: 10.1038/leu.2009.94. [DOI] [PubMed] [Google Scholar]
- 253.Brandwein J.M. Targeting polo-like kinase 1 in acute myeloid leukemia. Ther. Adv. Hematol. 2015;6:80–87. doi: 10.1177/2040620715571077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sistigu A., Yamazaki T., Vacchelli E., Chaba K., Enot D.P., Adam J., Vitale I., Goubar A., Baracco E.E., Remédios C., et al. Cancer cell–autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 2014;20:1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 255.Berneman Z.N., Anguille S., Van Marck V., Schroyens W.A., Van Tendeloo V.F. Induction of complete remission of acute myeloid leukaemia by pegylated interferon-α-2a in a patient with transformed primary myelofibrosis. Br. J. Haematol. 2010;149:152–155. doi: 10.1111/j.1365-2141.2009.08029.x. [DOI] [PubMed] [Google Scholar]
- 256.Anguille S., Lion E., Willemen Y., Van Tendeloo V.F.I., Berneman Z.N., Smits E.L.J.M. Interferon-α in acute myeloid leukemia: An old drug revisited. Leukemia. 2011;25:739–748. doi: 10.1038/leu.2010.324. [DOI] [PubMed] [Google Scholar]
- 257.Smits E.L.J.M., Anguille S., Berneman Z.N. Interferon α may be back on track to treat acute myeloid leukemia. Oncoimmunology. 2013;2:e23619. doi: 10.4161/onci.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Birkbak N.J., Eklund A.C., Li Q., McClelland S.E., Endesfelder D., Tan P., Tan I.B., Richardson A.L., Szallasi Z., Swanton C. Paradoxical Relationship between Chromosomal Instability and Survival Outcome in Cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zasadil L.M., Andersen K.A., Yeum D., Rocque G.B., Wilke L.G., Tevaarwerk A.J., Raines R.T., Burkard M.E., Weaver B.A. Cytotoxicity of Paclitaxel in Breast Cancer Is due to Chromosome Missegregation on Multipolar Spindles. Sci. Transl. Med. 2014;6:229ra43. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Jamal-Hanjani M., A’Hern R., Birkbak N.J., Gorman P., Grönroos E., Ngang S., Nicola P., Rahman L., Thanopoulou E., Kelly G., et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann. Oncol. 2015;26:1340–1346. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- 261.O’Neil N.J., Bailey M.L., Hieter P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 262.McBride A., Houtmann S., Wilde L., Vigil C., Eischen C.M., Kasner M., Palmisiano N. The Role of Inhibition of Apoptosis in Acute Leukemias and Myelodysplastic Syndrome. Front. Oncol. 2019;9 doi: 10.3389/fonc.2019.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Pan R., Ruvolo V., Mu H., Leverson J.D., Nichols G., Reed J.C., Konopleva M., Andreeff M. Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell. 2017;32:748–760.e6. doi: 10.1016/j.ccell.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gorodetska I., Kozeretska I., Dubrovska A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer. 2019;10:2109–2127. doi: 10.7150/jca.30410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Esposito M.T., So C.W.E. DNA damage accumulation and repair defects in acute myeloid leukemia: Implications for pathogenesis, disease progression, and chemotherapy resistance. Chromosoma. 2014;123:545–561. doi: 10.1007/s00412-014-0482-9. [DOI] [PubMed] [Google Scholar]
- 266.Alcalay M., Meani N., Gelmetti V., Fantozzi A., Fagioli M., Orleth A., Riganelli D., Sebastiani C., Cappelli E., Casciari C., et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Krejci O., Wunderlich M., Geiger H., Chou F.-S., Schleimer D., Jansen M., Andreassen P.R., Mulloy J.C. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111:2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Esposito M.T., Zhao L., Fung T.K., Rane J.K., Wilson A., Martin N., Gil J., Leung A.Y., Ashworth A., Eric So C.W. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat. Med. 2015;21:1481–1490. doi: 10.1038/nm.3993. [DOI] [PubMed] [Google Scholar]
- 269.Faraoni I., Compagnone M., Lavorgna S., Angelini D.F., Cencioni M.T., Piras E., Panetta P., Ottone T., Dolci S., Venditti A., et al. BRCA1, PARP1 and γH2AX in acute myeloid leukemia: Role as biomarkers of response to the PARP inhibitor olaparib. Biochim. Biophys. Acta Mol. Basis Dis. 2015;1852:462–472. doi: 10.1016/j.bbadis.2014.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.