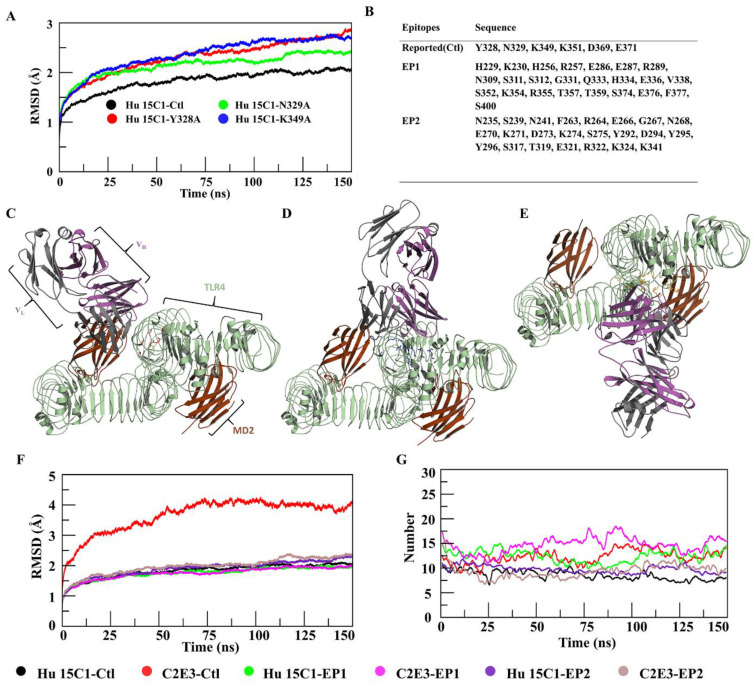
Figure 3.
Epitope prediction and docking mode and stability analysis. (A) RMSDs of four complexes in which mutated complexes showed an increasing trend. (B) Residues for the construction of three conformational epitopes. Modes of docking of Hu 15C1 with TLR4 at sites (C) Ctl, (D) EP1, and (E) EP2 with superimposition of the human TLR4–MD2 dimer. RMSDs of six complexes are illustrated in (F), where C2E3–Ctl (red) showed a maximum deviation, and all remaining complexes were stable. The number of hydrogen bonds during the simulation was determined in (G), where C2E3–EP1 (magenta) featured the highest, C2E3–Ctl (red) and Hu 15C1–EP1 (green) moderate, and Hu 15C1–Ctl (black), Hu 15C1–EP2 (blue), and C2E3–EP2 (brown) the lowest number of hydrogen bonds formed during the simulation.