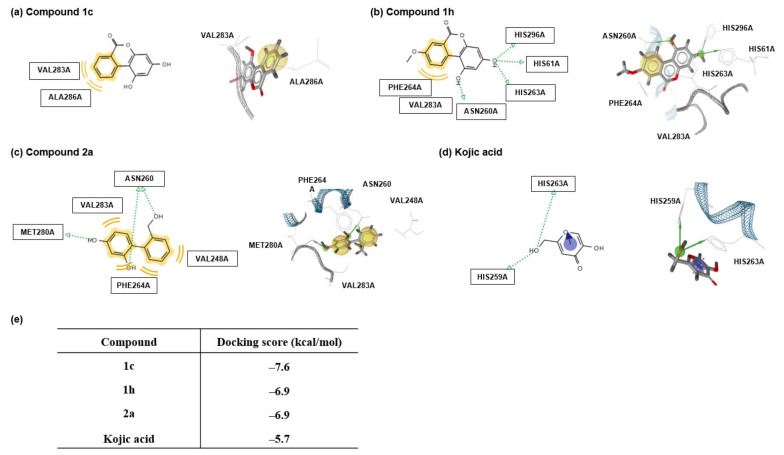
Figure 4.
Docking simulation of urolithin derivatives 1c and 1h, o-phenylbenzyl alcohol derivative 2a, and kojic acid with Agaricus bisporus tyrosinase using AutoDock Vina and pharmacophore analysis. (a–d) Pharmacophore results of 1c, 1h, 2a, and kojic acid obtained using LigandScout 4.2.1 based on AutoDock Vina indicated possible hydrophobic, π-π stacking, and hydrogen bonding interactions between tyrosinase amino acid residues and the ligands (shown in yellow and indicated by violet and green arrows, respectively). Docking simulation results showed hydrophobic (yellow spheres), π-π stacking (violet ring), and hydrogen bonding (green spheres) regions on ligands. (e) Docking scores of 1c, 1h, 2a, and kojic acid with tyrosinase are tabulated (PDB code: 2Y9X).