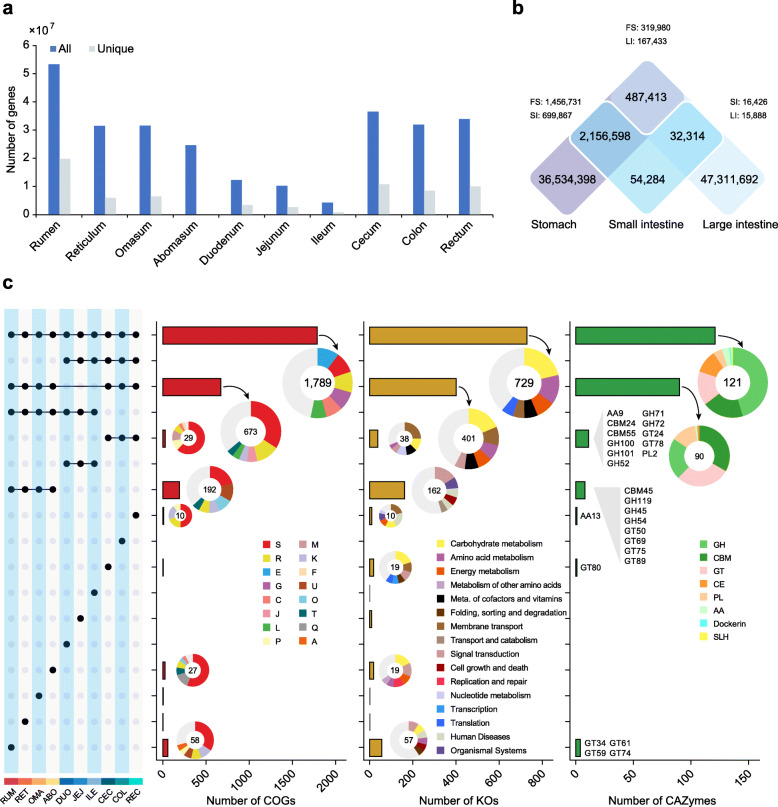
Fig. 3.
Specific functional features of the ruminant GIT microbiome. a All the nonredundant microbial genes across the ruminant GIT microbiome, with unique counts in each region. b Venn diagram of the unique and shared gene counts between the stomach (FS: rumen, reticulum, omasum, and abomasum), small intestine (SI: duodenum, jejunum, and ileum), and large intestine (LI: cecum, colon, and rectum). c Comparison of the levels of functional modules (COGs, KOs, and CAZymes) of the microbiome across regions of the ruminant GIT. The left panel shows sets included in the intersection and independent sites, and the right bar or pie charts show the categories of the functional modules in these sets. The major enriched categories are shown in the legend. A, RNA processing and modification; C, energy production and conversion; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport. GH, glycoside hydrolases; GT, glycosyl transferases; CE, carbohydrate esterases; PL, polysaccharide lyases; CBM, carbohydrate-binding modules; SLH, S-layer homology module; AA, auxiliary activities