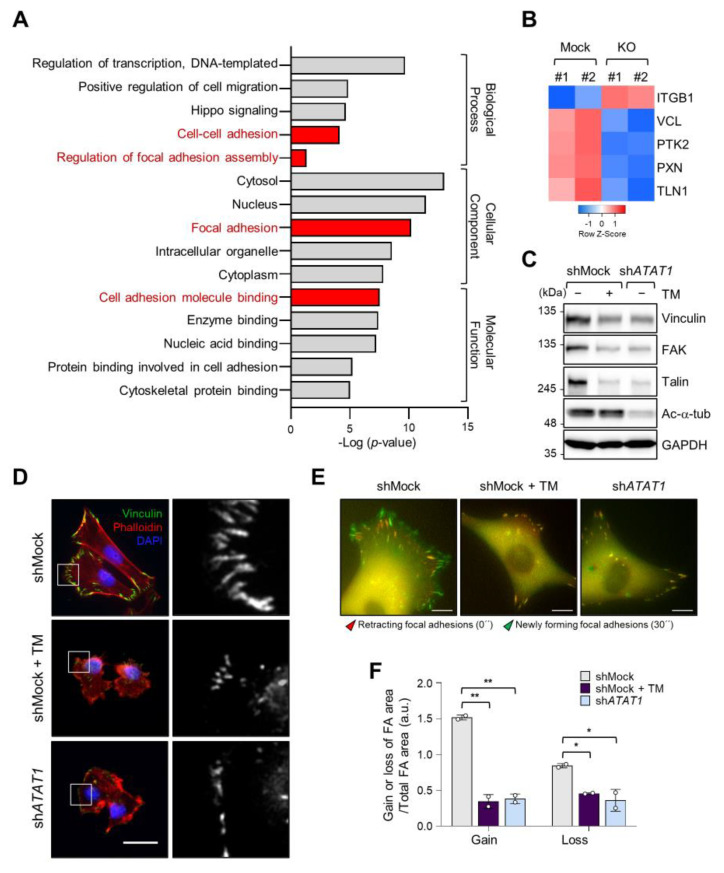
Figure 4.
Focal adhesion assembly is regulated by microtubule acetylation and ER stress. (A) Functional annotation of 389 DEGs in ATAT1 KO MDA-MB-231 cells using PANTHER gene ontology (GO). (B) Heatmap showing genes related to focal adhesion assembly based on RNA-seq data from ATAT1 KO cells. (C) Validation of gene expression using Western blotting. shMock-treated cells treated or not with tunicamycin, and shATAT1-treated cells were cultured for 24 h. Cell lysates were used for Western blot analysis of vinculin, FAK, talin, and acetylated α-tubulin, using GAPDH as a loading control. (D) Immunocytochemistry analysis of focal adhesions using antibodies against vinculin and F-actin in shMock-treated and ATAT1 KO cells cultured in the presence of 20 ng/mL tunicamycin for 24 h. Scale bar, 30 μm. (E) Paxillin-GFP-expressing shMock- and shATAT1 #1-treated MDA-MB-231 cells were starved for 16 h in serum-free RPMI1640 medium and then stimulated with 10% FBS with or without 20 ng/mL tunicamycin. Merged paxillin–GFP images in shMock- and shATAT1 #1-treated cells at 0 and 30 min. Red represents retracting focal adhesions and green represents newly forming focal adhesions. Scale bar, 10 μm. (F) Ratios of gain and loss of focal adhesions to total focal adhesion area. Values represent the mean ± SD of two independent experiments. Statistical significances were analyzed using one-way ANOVA followed by Tukey multiple comparison tests. One-way ANOVA (Loss; F2, 3 = 15.98. * p < 0.05, Gain; F2, 3 = 174.1, ** p < 0.01).