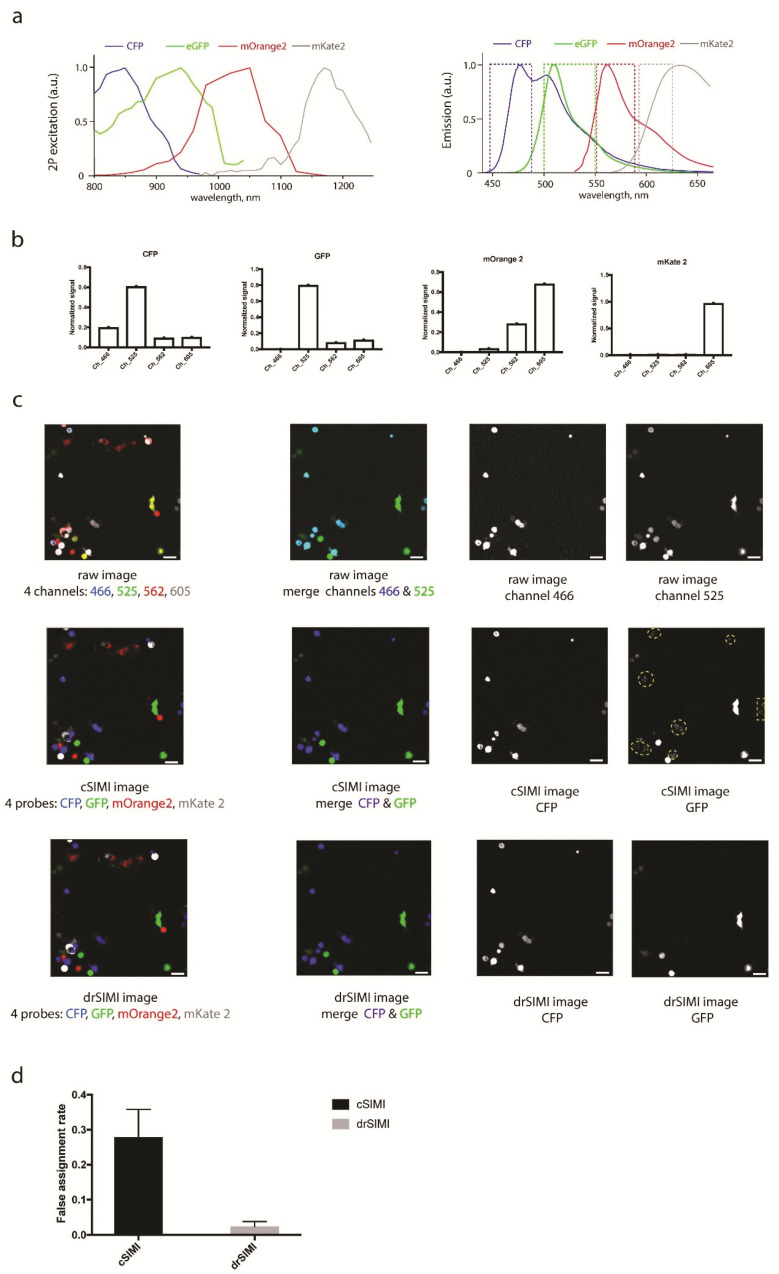
Figure 1.
(a) Two-photon excitation and emission spectra of cyan fluorescent protein (CFP), enhanced green fluorescent protein (eGFP), a monomeric orange fluorescent protein (mOrange2), and a monomeric red fluorescent protein (mKate2). (b) Spectral signatures (fingerprints) of CFP, eGFP, mOrange2, and mKate2 used for both drSIMI and cSIMI approaches. (c) Two-photon fluorescence images of mixed HEK cells transfected with CFP, GFP, mOrange2, or mKate2, together with the results of cSIMI and drSIMI. First column: merged fluorescence images containing all four probes; second column: merged fluorescence images containing only the CFP and eGFP probes, i.e., the probes that represent a spectral unmixing challenge; third column: CFP image layer; fourth column: eGFP image layer. The yellow dashed-line areas depict a crosstalk pattern of the CFP probes into the GFP image layer. (d) Quantification of the unmixing accuracy (false assignment rate of the chromophore within a segmented object) of the pixel-based similarity unmixing approaches (0—no false assignment; 1—no correct assignment). Scale bar, 50 μm.