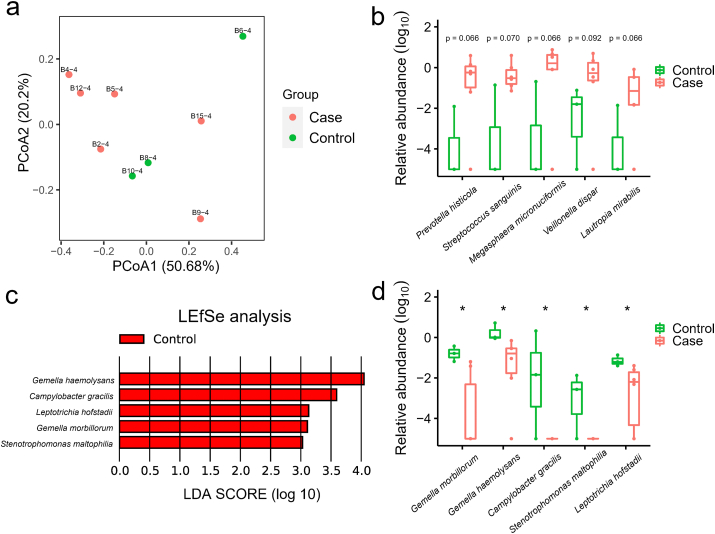
Fig. 3.
The metagenome analysis of the nasopharyngeal microbiome in COVID-19 and non- COVID-19 patients. (a) PCoA plot shows the composition of nasopharyngeal microbiota at the family level in COVID-19 (Case, n = 6) and non-COVID-19 (Control, n = 3) patients. (b) The species that were more abundant in the nasopharyngeal samples of COVID-19 patients with p value less than 0.1 (but not less than 0.05). The mean values and error bars were shown in the boxplots. P values, Wilcoxon rank-sum test, of each species were labelled above the boxplots. (c) The LDA Effect Size (LEfSe) analysis of the species in relative abundance between COVID-19 (Case) and non-COVID-19 (Control) patients. Five species were enriched in non-COVID-19 (Control) patients according to the LDA scores. (d) The significantly different species in relative abundance of nasopharyngeal microbiome between COVID-19 (Case) and non-COVID-19 (Control) patients. *, p value < 0.05.