Figure 1.
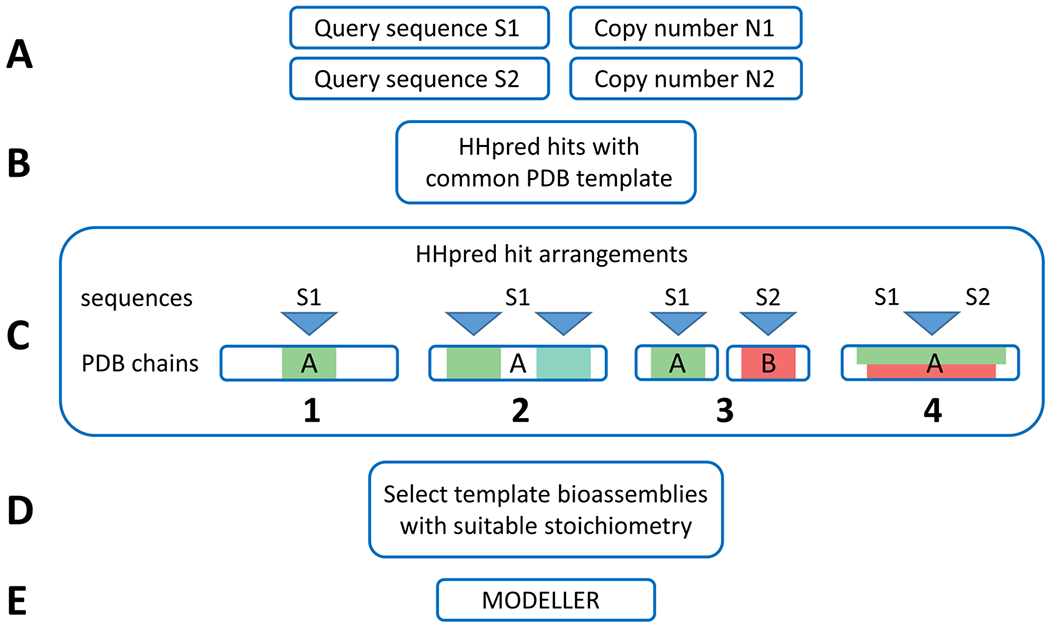
General outline of the ClusPro template-based modeling (TBM) protocol. (A) ClusPro TBM takes component sequences and their corresponding copy numbers in the modeled assembly as inputs. (B) HHpred is used to find potential templates for each query sequence, and HHpred hits sharing a common structural template are identified. (C) The HHpred hits are combined to obtain potential assembly-generating arrangements for each template structure (arrangements in the figure are examples, and are not necessarily generated as a part of a single server job). (D) The arrangements are evaluated on their ability to produce a model with user-specified stoichiometry based on biological assemblies specified in the template PDB file. (E) Arrangements passing the stoichiometry filter are used to construct the assembly models using MODELLER.
