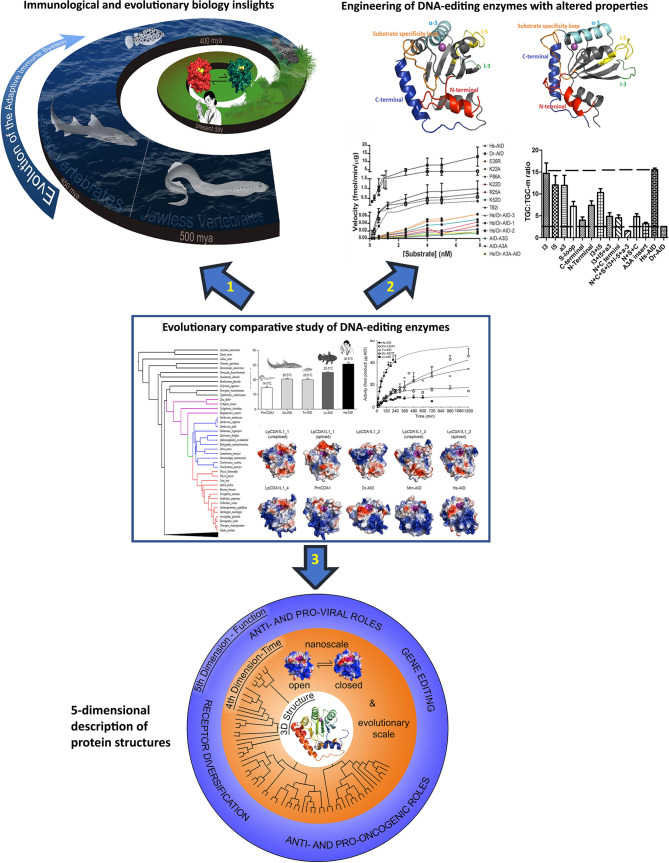
Figure 2.
The concept and three main applications of the evolutionary-biochemical-computational approach to studying DNA-editing enzymes. The evolutionary comparative approach is shown in the middle with 3 arrows each pointing to an area wherein this approach can make significant impact. The evolutionary comparative approach shown in the middle panel consists of comparing biochemical properties (Michaelis-Menten kinetics, substrate binding kinetics, optimal temperature, optimal pH, substrate sequence or shape specificity, etc.) of the enzymes using enzyme assays and considering insights in the context of their 3D solved structures or computational predicted models as shown in this figure. Due to vast biochemical diversity observed amongst various AID orthologs, examining the biochemical properties of divergent AID orthologs has shed light on many structure:function aspects of AID/APOBEC enzymes. Arrow 1: the evolutionary comparative study of DNA-editing enzymes can provide insights into the evolution of the immune system, for instance on whether the immune systems use active deaminases and how/if they have gene sequences or other immune genes that have co-evolved with their deaminases. Arrow 2: using different orthologs allows for generation of libraries of mutants and chimeric enzymes which can have diverse biochemical properties such as DNA/RNA-targeting profiles and sequence specificities, and these can be used for applications such as base editing. Arrow 3: the most important highlight of the evolutionary-biochemical-computational approach is the birth of the concept of 5-dimentional (5D) structural description, proposed in this article. The 5D description integrates the classical 3D structure of a protein with dynamic changes in time (4th dimension) and the relevance of these to function (5th dimension). The middle panel contains reproduced figures from previous publications. The thermosensitivity and enzyme velocity plots are from our previous work Quinlan EM et al. (21). Biochemical regulatory features of activation-induced cytidine deaminase remain conserved from lampreys to humans. Mol Cell Biol 37:e00077-17. https://doi.org/10.1128/MCB.00077-17. Copyright © 2017 American Society for Microbiology. The computational models are adapted from our previous work Holland et al. (20). Expansions, diversification, and interindividual copy number variations of AID/APOBEC family cytidine deaminase genes in lampreys. 2018 Apr 3;115(14):E3211-E3220. doi: 10.1073/pnas.1720871115 Copyright (2018) National Academy of Sciences.