Table 1.
Exogenous sources of ICLs, their preferential DNA targets, and representative chemical structures for each.
| ICL Agent | Target Sequence | Structurea |
|---|---|---|
| Mitomycin C | 5′-CG | 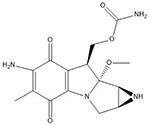 |
| Pyrrolobenzodiazepines | Purine-GATC-pyrimidine | 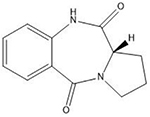 |
| Colibactin | 5′-AATTT and 5′-ATTTT | 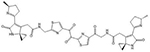 |
| Psoralens | 5′-TA | 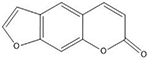 |
| Pyrrolizidine alkaloids | N2 guanine in 5′-CG-3′ | 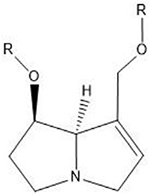 |
| Nitrogen mustards | 5′-GNC | |
| Chloroethylnitrosoureas | O6- and N7 positions of guanine | 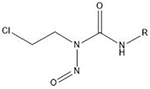 |
| Cisplatin | 5′-GC | 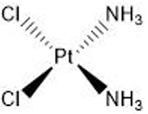 |
| Diepoxybutane | 5′-GNC | 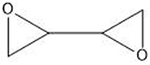 |
| Dianhydrogalactitol | N7 guanine | 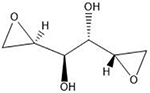 |
Many of these structures can vary by the presence of a number of different possible–R groups. For the sake of brevity, the simplest molecular scaffolds are shown.
