Table 1.
Receptor-Binding Profiles of Truncated 1′-Homologated Adenosine Derivatives 4a–4t
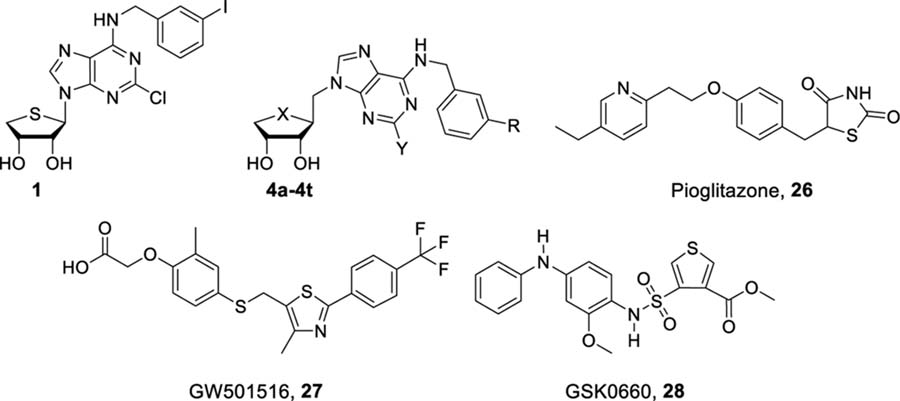 | |||||||
|---|---|---|---|---|---|---|---|
| cpd | X | Y | R | hA3AR Ki (nM) or % replacementa | PPARα % replacementb at 30 μM (%) | PPARγ Ki (μM) or % replacementb | PPARδ Ki (μM) or % replacementb |
| 1 | 4.16 | 35.6 | 3.33 ± 1.67 | 0.0106 ± 0.015 | |||
| 4a | S | Cl | F | 17% at 10 μM | 15.5 | 3.89 ± 0.47 | 1.91 ± 0.39 |
| 4b | S | Cl | Cl | 13% at 10 μM | 12.8 | 34.1% at 30 μM | 7.8% at 30 μM |
| 4c | S | Cl | Br | 21% at 10 μM | 3.0 | 4.07 ± 0.86 | 0.592 ± 0.26 |
| 4d | S | Cl | I | 10% at 10 μM | 20.4 | 1.23 ± 0.25 | 0.106 ± 0.031 |
| 4e | S | H | F | 9% at 10 μM | 25.6 | 24.4% at 30 μM | 0.8% at 30 μM |
| 4f | S | H | Cl | 9% at 10 μM | 7.7 | 24.9% at 30 μM | 10.0% at 30 μM |
| 4g | S | H | Br | 6% at 10 μM | 5.4 | 33.5% at 30 μM | 9.9% at 30 μM |
| 4h | S | H | I | 14% at 10 μM | 9.4 | 39.2% at 30 μM | 5.2% at 30 μM |
| 4i | S | −C≡CCH3 | F | 2% at 10 μM | 15.5 | 7.66 ± 0.65 | 13.8% at 30 μM |
| 4j | S | −C≡CCH3 | Cl | 1% at 10 μM | 12.6 | 41.7% at 30 μM | 3.8% at 30 μM |
| 4k | S | −C≡CCH3 | Br | 12% at 10 μM | 12.8 | 7.04 ± 0.40 | 15.3% at 30 μM |
| 4l | S | −C≡CCH3 | I | 2% at 10 μM | 24.0 | 7.30 ± 1.01 | 15.9% at 30 μM |
| 4m | O | Cl | F | 2% at 10 μM | 5.0 | 9.0% at 30 μM | 7.3% at 30 μM |
| 4n | O | Cl | Cl | 1% at 10 μM | 3.6 | 7.66 ± 2.45 | 0.504 ± 0.29 |
| 4o | O | Cl | Br | 14% at 10 μM | 23.2 | 9.55 ± 6.61 | 1.43 ± 0.67 |
| 4p | O | Cl | I | 3% at 10 μM | 0.3 | 3.56 ± 0.75 | 0.374 ± 0.036 |
| 4q | O | H | F | 1% at 10 μM | 1.4 | 13.3% at 30 μM | 14.6% at 30 μM |
| 4r | O | H | Cl | 1% at 10 μM | 8.3 | 0.8% at 30 μM | 10.6% at 30 μM |
| 4s | O | H | Br | 14% at 10 μM | 5.6 | 4.1% at 30 μM | 16.5% at 30 μM |
| 4t | O | H | I | 3% at 10 μM | 1.7 | 16.0% at 30 μM | 6.5% at 30 μM |
| 26 | ND | 3.5 | 0.096 ± 0.049 | 14.7% at 30 μM | |||
| 27 | ND | 44.9 | 38.5% at 30 μM | 0.00608 ± 0.0055 | |||
| 28 | ND | 12.4 | 59.9% at 30 μM | 0.00465 ± 0.0023 | |||
